Robustness Analyses
Here, we perform additional robustness checks including age in each model as a covariate.
Library
## Lade nötiges Paket: carData## lattice theme set by effectsTheme()
## See ?effectsTheme for details.## Lade nötiges Paket: Matrix##
## Attache Paket: 'sjstats'## Die folgenden Objekte sind maskiert von 'package:effectsize':
##
## cohens_f, cramers_v, phi##
## Attache Paket: 'lmerTest'## Das folgende Objekt ist maskiert 'package:lme4':
##
## lmer## Das folgende Objekt ist maskiert 'package:stats':
##
## step##
## Attache Paket: 'dplyr'## Die folgenden Objekte sind maskiert von 'package:formr':
##
## first, last## Die folgenden Objekte sind maskiert von 'package:stats':
##
## filter, lag## Die folgenden Objekte sind maskiert von 'package:base':
##
## intersect, setdiff, setequal, unionData
Load selected data based on 03_codebook
Political, Ethnic, and Religious Similarity
H1a Preference for Similarity in Political Beliefs
H1a(1) There is no linear link between right-wing political orientation and women’s preferences for partner’s similar political beliefs and values. H1a(2) There is a positive quadratic link between right-wing political orientation and women’s preferences for partner’s similar political beliefs and values. Outcome: Preference ratings for partner’s similar political beliefs and values. Predictors: Political Orientation & Age. Random intercept and random slope for country.
H1a(1) Linear Effect
Models
Summary
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula:
## pref_politicalsim ~ political_orientation + age + (1 + political_orientation |
## country)
## Data: data_included_documented
## Control: lmerControl(optimizer = "bobyqa")
##
## REML criterion at convergence: 52404.8
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.5217 -0.6811 0.1268 0.7383 2.2603
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## country (Intercept) 0.7130 0.8444
## political_orientation 0.0326 0.1806 -0.83
## Residual 3.3023 1.8172
## Number of obs: 12946, groups: country, 144
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 2.692e+00 1.338e-01 1.049e+02 20.116 <2e-16 ***
## political_orientation -7.417e-02 3.175e-02 4.687e+01 -2.336 0.0238 *
## age 2.853e-02 2.356e-03 1.291e+04 12.112 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) pltcl_
## pltcl_rnttn -0.759
## age -0.439 0.021## Computing profile confidence intervals ...## 0.15 % 99.85 %
## .sig01 0.56024595 1.22559147
## .sig02 -0.95021383 -0.46510688
## .sig03 0.10187680 0.29165818
## .sigma 1.78396082 1.85138428
## (Intercept) 2.27776577 3.09149283
## political_orientation -0.16911363 0.02952843
## age 0.02153767 0.03552314Standardized Coefficients
## # A tibble: 3 × 5
## Parameter Std_Coefficient CI CI_low CI_high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0 0.997 0 0
## 2 political_orientation -0.0528 0.997 -0.120 0.0143
## 3 age 0.103 0.997 0.0775 0.128Plot
lmer(pref_politicalsim ~ political_orientation + age + (1+political_orientation|country),
data = data_included_documented) %>%
allEffects() %>%
plot()## Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
## Model failed to converge with max|grad| = 0.00397324 (tol = 0.002, component 1)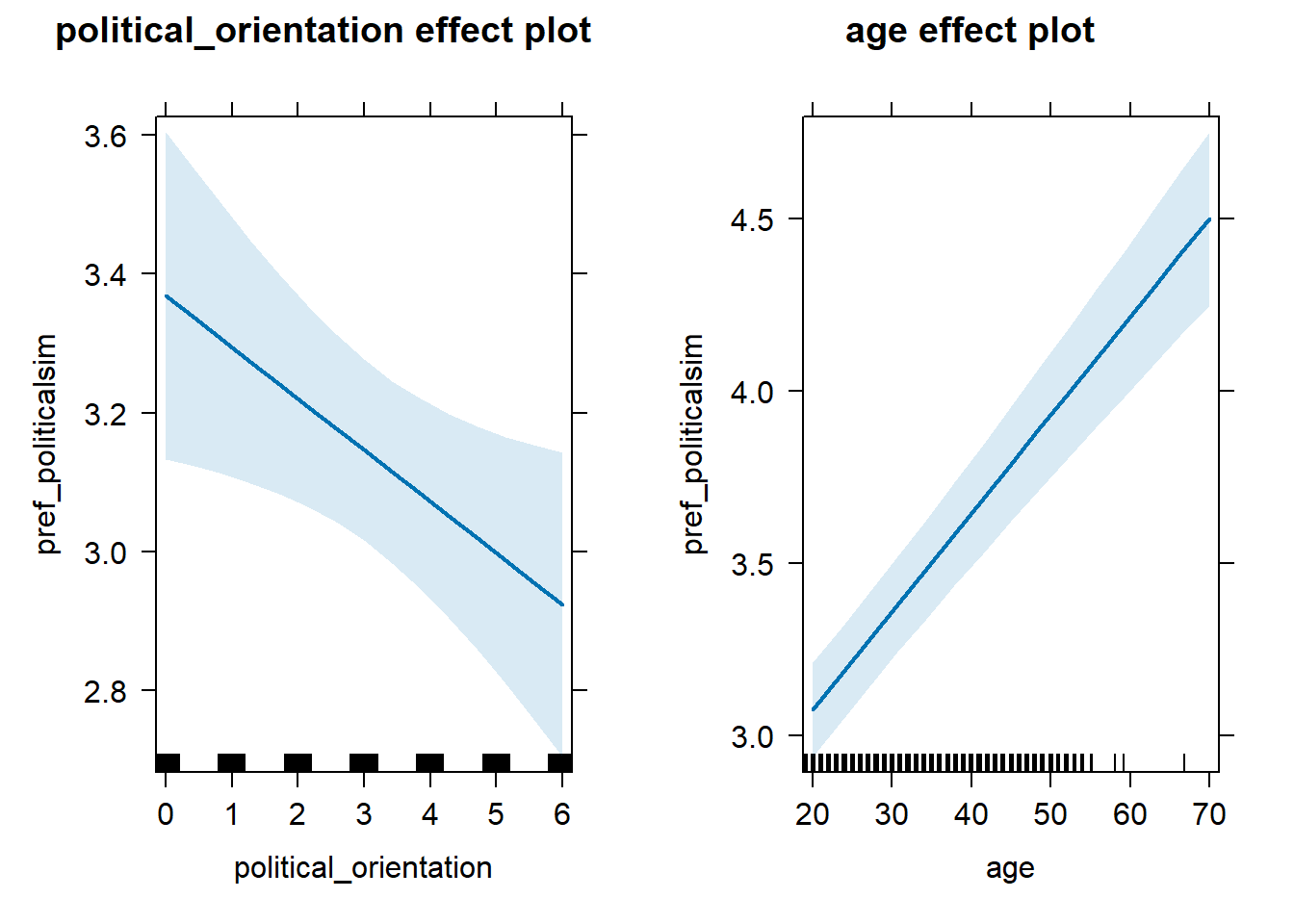
H1a(2) Quadratic Effect
Here, we are examining the quadratic effect of right-wing political orientation on preferred political similarity in a partner controlling for age using the Two Lines Approach (Simonsohn, 2018). We are using the Robin Hood Algorithm in order to set the breaking point. Then, we are calculating two multilevel regressions on either side of the breaking point. Outcome: Preference ratings for partner’s similar political beliefs and values. Predictors: Political Orientation & Age. Random intercept and random slope for country.
Algorithm: Find breaking point
See 11_twolines_analyses_multilevel
Regression 1 (x <= breaking_point)
model_pref_politicalsim_1_robustcheck = lmer(pref_politicalsim ~ political_orientation + age +
(1+political_orientation|country),
data = data_included_documented %>%
dplyr::filter(political_orientation <= 3), control =lmerControl(optimizer = "bobyqa"))
summary(model_pref_politicalsim_1_robustcheck)## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula:
## pref_politicalsim ~ political_orientation + age + (1 + political_orientation |
## country)
## Data: data_included_documented %>% dplyr::filter(political_orientation <=
## 3)
## Control: lmerControl(optimizer = "bobyqa")
##
## REML criterion at convergence: 42551.7
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.7593 -0.6977 0.1446 0.7186 2.3337
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## country (Intercept) 0.62089 0.7880
## political_orientation 0.05005 0.2237 -0.74
## Residual 3.14588 1.7737
## Number of obs: 10633, groups: country, 138
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 3.135e+00 1.383e-01 7.205e+01 22.666 <2e-16 ***
## political_orientation -3.030e-01 4.200e-02 2.907e+01 -7.214 6e-08 ***
## age 2.756e-02 2.559e-03 1.060e+04 10.767 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) pltcl_
## pltcl_rnttn -0.730
## age -0.463 0.021## Computing profile confidence intervals ...## 0.15 % 99.85 %
## .sig01 0.46632087 1.2306668
## .sig02 -0.92602352 -0.1616867
## .sig03 0.09393412 0.4034475
## .sigma 1.73786214 1.8106810
## (Intercept) 2.67745475 3.5563482
## political_orientation -0.42985824 -0.1479782
## age 0.01996013 0.0351559## # A tibble: 3 × 5
## Parameter Std_Coefficient CI CI_low CI_high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0 0.997 0 0
## 2 political_orientation -0.164 0.997 -0.231 -0.0965
## 3 age 0.0984 0.997 0.0713 0.126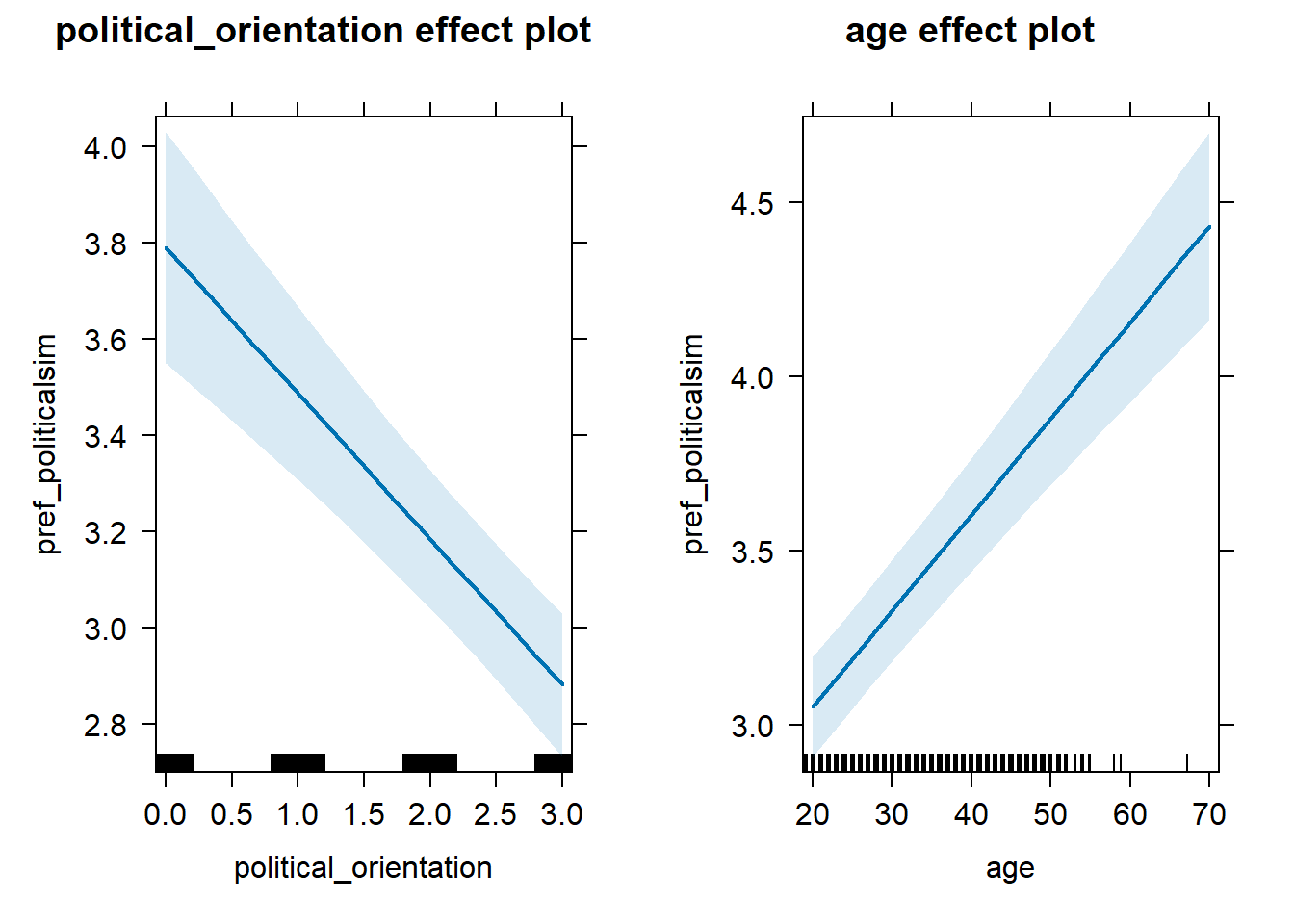
Regression 2 (x >= breaking_point)
model_pref_politicalsim_2_robustcheck = lmer(pref_politicalsim ~ political_orientation + age +
(1+political_orientation|country),
data = data_included_documented %>%
dplyr::filter(political_orientation >= 3), control =lmerControl(optimizer = "bobyqa"))
summary(model_pref_politicalsim_2_robustcheck)## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula:
## pref_politicalsim ~ political_orientation + age + (1 + political_orientation |
## country)
## Data: data_included_documented %>% dplyr::filter(political_orientation >=
## 3)
## Control: lmerControl(optimizer = "bobyqa")
##
## REML criterion at convergence: 29754.8
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -3.0355 -0.8181 0.1015 0.7168 2.2669
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## country (Intercept) 0.7411 0.8609
## political_orientation 0.0311 0.1763 -0.81
## Residual 3.2812 1.8114
## Number of obs: 7356, groups: country, 129
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 9.904e-01 1.925e-01 3.369e+01 5.144 1.14e-05 ***
## political_orientation 4.090e-01 4.546e-02 2.269e+01 8.996 6.10e-09 ***
## age 2.643e-02 3.137e-03 7.337e+03 8.425 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) pltcl_
## pltcl_rnttn -0.838
## age -0.392 0.002## Computing profile confidence intervals ...## 0.15 % 99.85 %
## .sig01 0.31552709 1.64154220
## .sig02 -0.97162975 0.91451883
## .sig03 0.03947407 0.37782267
## .sigma 1.76753258 1.85696350
## (Intercept) 0.41240088 1.68463907
## political_orientation 0.23621054 0.55393090
## age 0.01711562 0.03573698## # A tibble: 3 × 5
## Parameter Std_Coefficient CI CI_low CI_high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0 0.997 0 0
## 2 political_orientation 0.175 0.997 0.117 0.233
## 3 age 0.0943 0.997 0.0611 0.128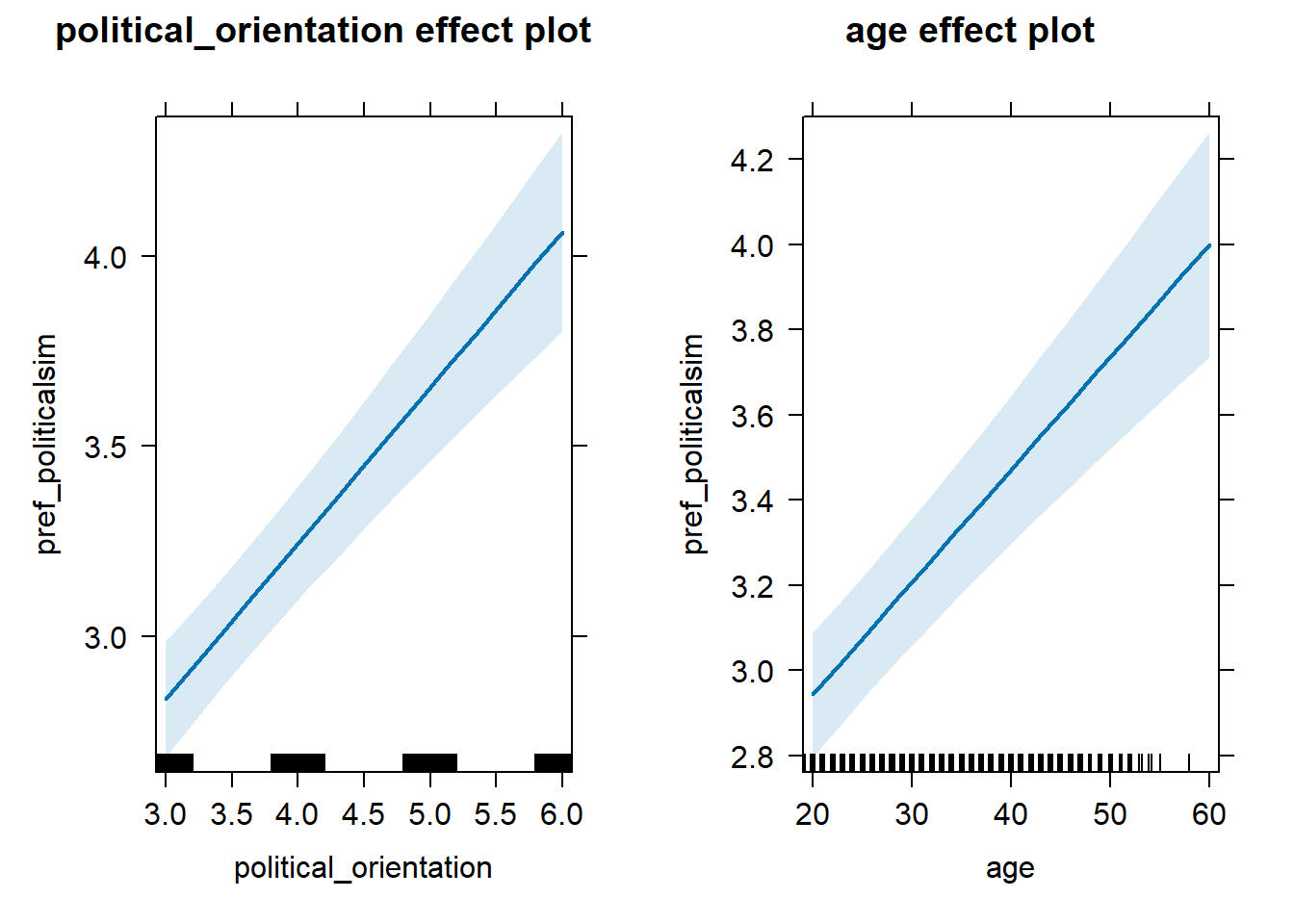
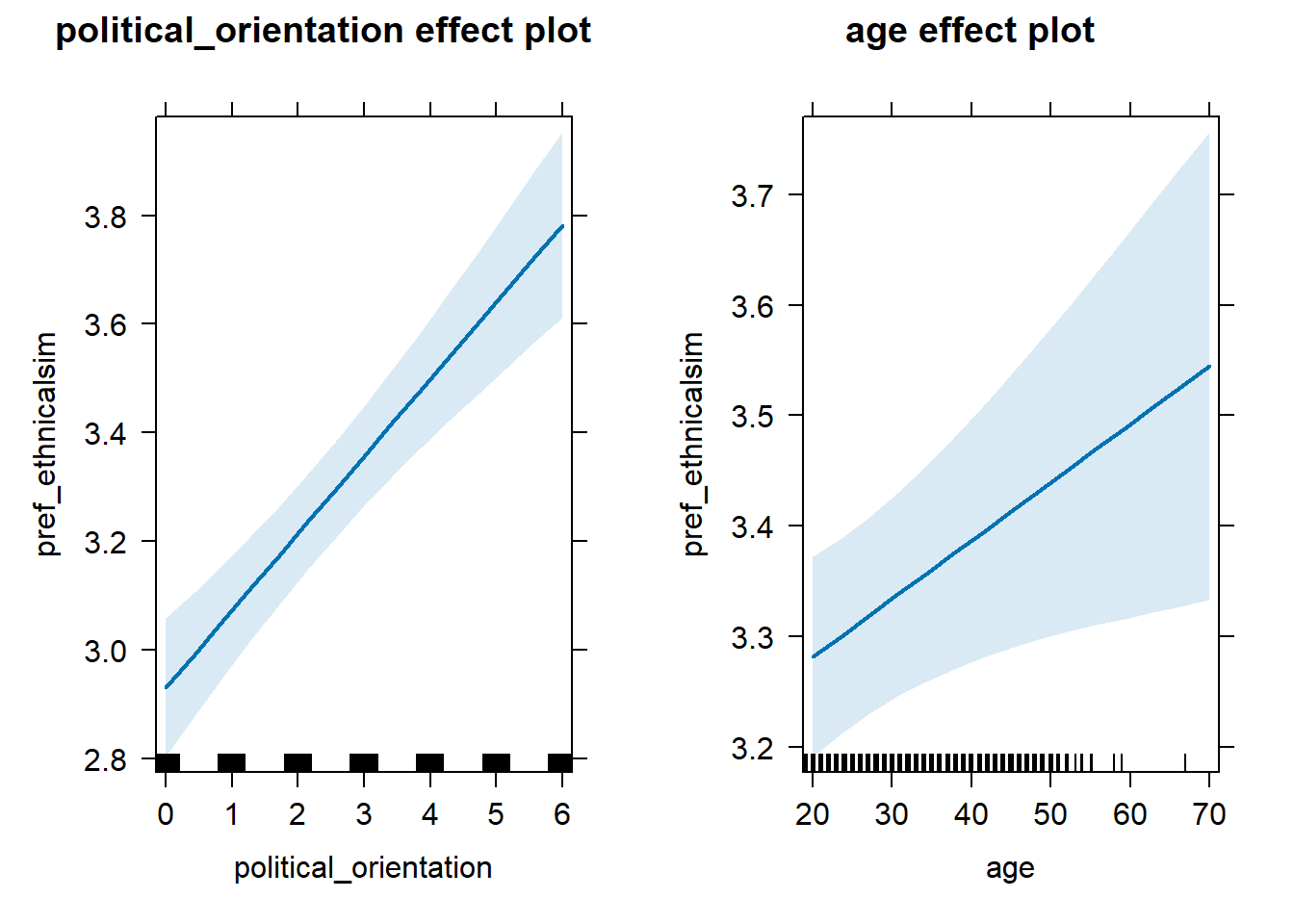
H1b Preference for Similarity in Ethnicity/Race
H1b There is a positive linear link between right-wing political orientation and women’s preferences for partner’s similar ethnicity/race. Outcome: Preference ratings for partner’s similar ethnicity/race. Predictors: Political Orientation & Age. Random intercept and random slope for country.
Models
Summary
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula:
## pref_ethnicalsim ~ political_orientation + age + (1 + political_orientation |
## country)
## Data: data_included_documented
## Control: lmerControl(optimizer = "bobyqa")
##
## REML criterion at convergence: 30430.5
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -3.2779 -0.4648 -0.0409 0.6023 2.4234
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## country (Intercept) 0.080758 0.28418
## political_orientation 0.006377 0.07986 -0.40
## Residual 1.955411 1.39836
## Number of obs: 8641, groups: country, 127
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 2.805e+00 8.451e-02 6.408e+01 33.188 < 2e-16 ***
## political_orientation 1.420e-01 2.040e-02 2.529e+01 6.962 2.52e-07 ***
## age 5.262e-03 2.158e-03 8.636e+03 2.438 0.0148 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) pltcl_
## pltcl_rnttn -0.572
## age -0.637 0.018## Computing profile confidence intervals ...## 0.15 % 99.85 %
## .sig01 0.104270231 0.5667020
## .sig02 -0.879729688 0.8025576
## .sig03 0.030113496 0.1646519
## .sigma 1.367138420 1.4306733
## (Intercept) 2.553361718 3.0755480
## political_orientation 0.071592573 0.2033169
## age -0.001137014 0.0116801Standardized Coefficients
## # A tibble: 3 × 5
## Parameter Std_Coefficient CI CI_low CI_high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0 0.997 0 0
## 2 political_orientation 0.129 0.997 0.0742 0.184
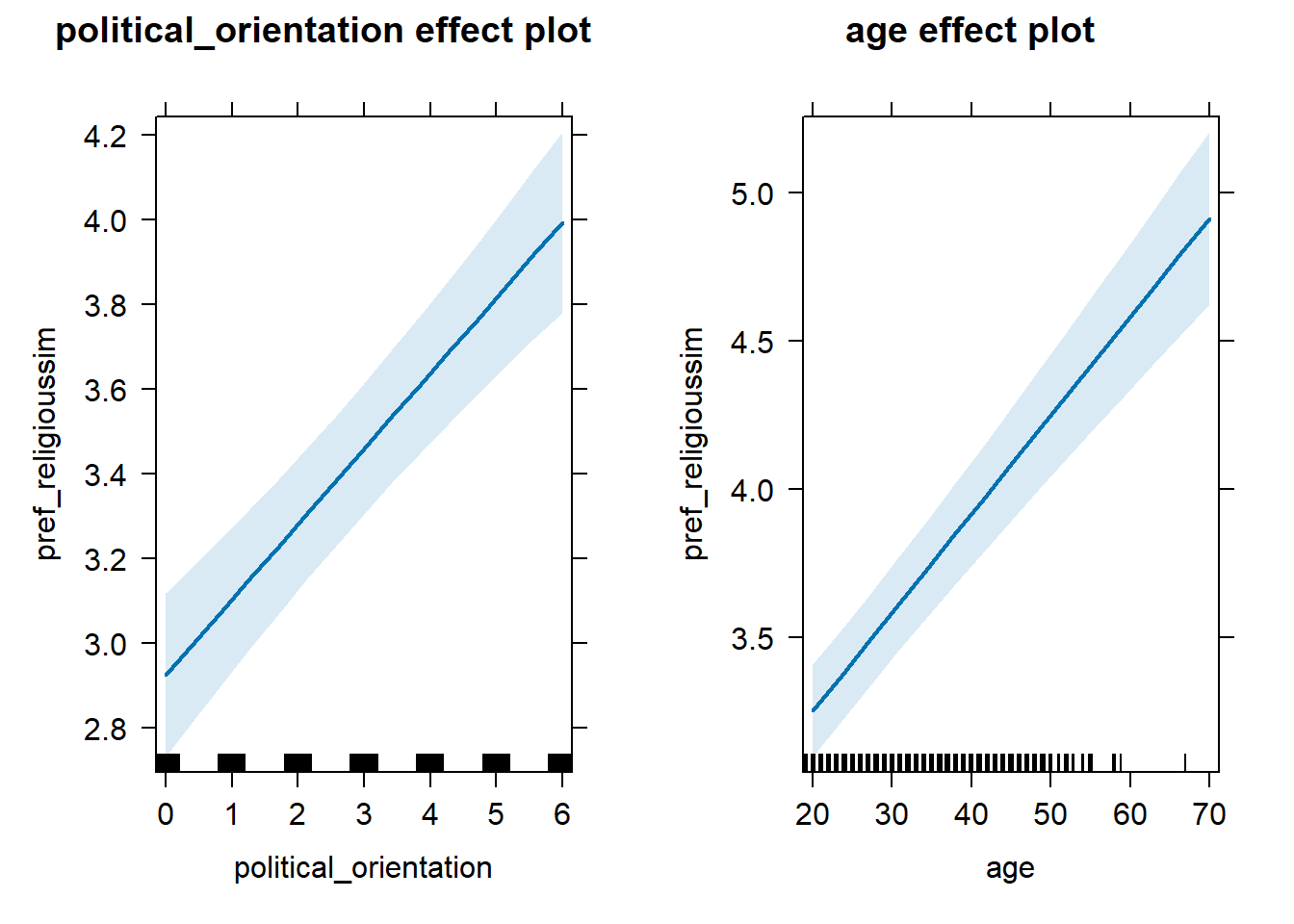
## 3 age 0.0258 0.997 -0.00562 0.0573H1c Preference for Similarity in Religion
H1c There is a positive linear link between right-wing political
orientation and women’s preferences for partner’s similar religious
beliefs.
Outcome: Preference ratings for partner’s similar religious beliefs.
Predictors: Political Orientation & Age. Random intercept and random
slope for country.
Models
Summary
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula:
## pref_religioussim ~ political_orientation + age + (1 + political_orientation |
## country)
## Data: data_included_documented
## Control: lmerControl(optimizer = "bobyqa")
##
## REML criterion at convergence: 56373.6
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.44074 -1.00588 0.05918 0.84691 2.04253
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## country (Intercept) 0.403206 0.63499
## political_orientation 0.006292 0.07932 -0.36
## Residual 4.519047 2.12581
## Number of obs: 12934, groups: country, 144
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 2.137e+00 1.200e-01 6.933e+01 17.803 < 2e-16 ***
## political_orientation 1.781e-01 2.255e-02 2.561e+01 7.895 2.52e-08 ***
## age 3.327e-02 2.751e-03 1.291e+04 12.094 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) pltcl_
## pltcl_rnttn -0.518
## age -0.570 0.033## Computing profile confidence intervals ...## 0.15 % 99.85 %
## .sig01 0.33549165 1.04178607
## .sig02 -0.91036705 0.94115335
## .sig03 0.02031065 0.17154676
## .sigma 2.08691097 2.16580486
## (Intercept) 1.75400882 2.57186768
## political_orientation 0.07661313 0.26975566
## age 0.02508655 0.04142527Standardized Coefficients
## # A tibble: 3 × 5
## Parameter Std_Coefficient CI CI_low CI_high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0 0.997 0 0
## 2 political_orientation 0.110 0.997 0.0688 0.152
## 3 age 0.104 0.997 0.0787 0.130Ideal Partner Preferences
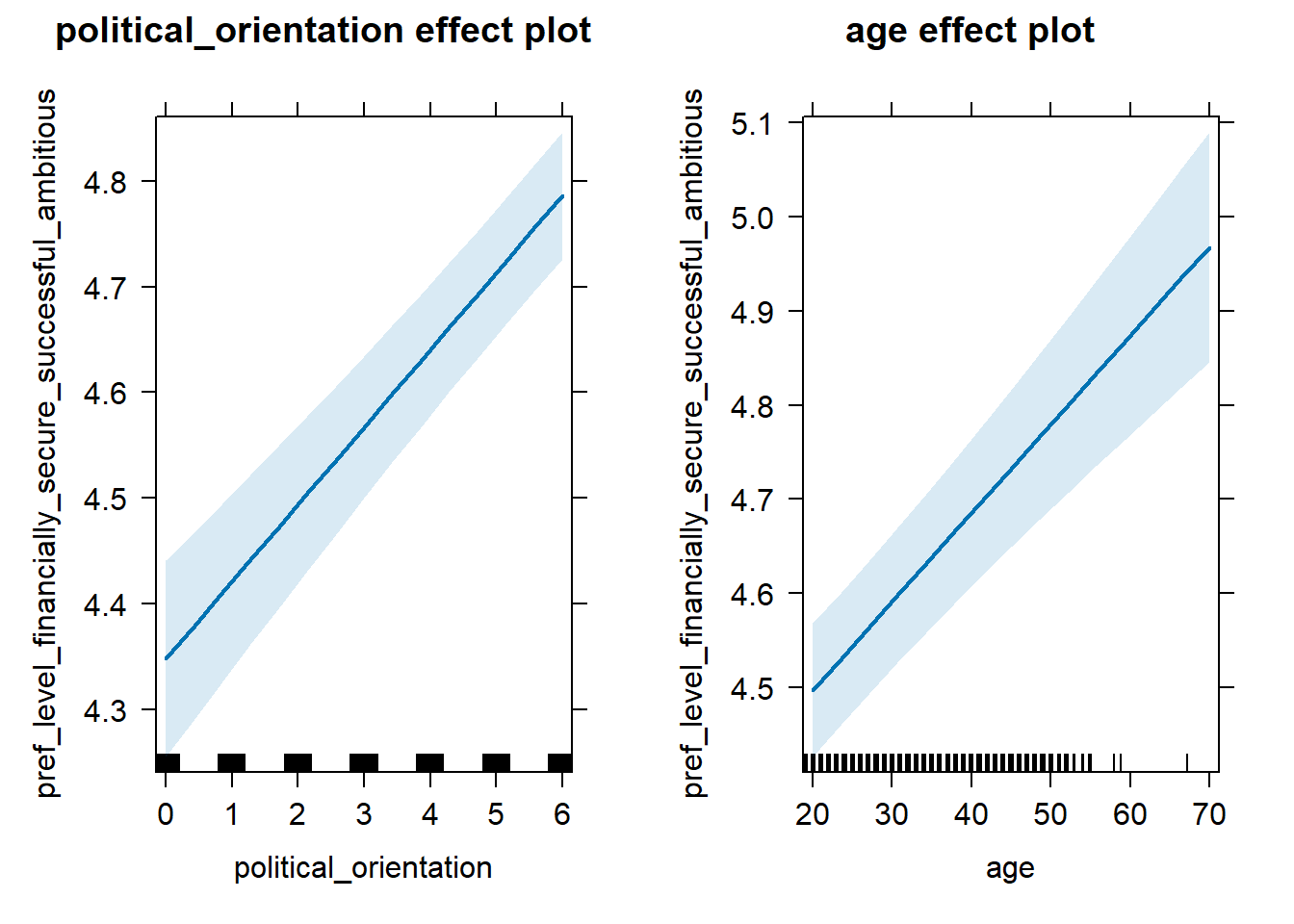
H2a Preference for the Level of Financial Security- Successfulness
H2a There is a positive linear link between right-wing political orientation and women’s preferences for the level of financial security and successfulness. Outcome: Level ratings for partner’s financial security-successfulness. Predictors: Political Orientation & Age. Random intercept and random slope for country.
Models
model_pref_level_financially_secure_successful_ambitious_robustcheck <- lmer(pref_level_financially_secure_successful_ambitious ~ political_orientation + age + (1+political_orientation|country), data = data_included_documented, control =lmerControl(optimizer = "bobyqa"))## boundary (singular) fit: see help('isSingular')Summary
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula:
## pref_level_financially_secure_successful_ambitious ~ political_orientation +
## age + (1 + political_orientation | country)
## Data: data_included_documented
## Control: lmerControl(optimizer = "bobyqa")
##
## REML criterion at convergence: 31651.5
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -4.8169 -0.6960 0.0004 0.6958 2.6418
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## country (Intercept) 0.1341548 0.36627
## political_orientation 0.0009063 0.03011 -1.00
## Residual 0.7195492 0.84826
## Number of obs: 12548, groups: country, 143
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 4.125e+00 5.499e-02 1.352e+02 75.004 <2e-16 ***
## political_orientation 7.306e-02 6.808e-03 9.832e+01 10.731 <2e-16 ***
## age 9.398e-03 1.114e-03 1.251e+04 8.439 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) pltcl_
## pltcl_rnttn -0.689
## age -0.509 0.056
## optimizer (bobyqa) convergence code: 0 (OK)
## boundary (singular) fit: see help('isSingular')## Computing profile confidence intervals ...## Warning in FUN(X[[i]], ...): non-monotonic profile for .sig02## Warning in confint.thpr(pp, level = level, zeta = zeta): bad spline fit for
## .sig02: falling back to linear interpolation## 0.15 % 99.85 %
## .sig01 0.253369012 0.52378807
## .sig02 -1.006492439 -0.63548979
## .sig03 0.008973381 0.05673334
## .sigma 0.835930822 0.85796383
## (Intercept) 3.960542261 4.30049507
## political_orientation 0.047481689 0.09489592
## age 0.006092425 0.01270256Standardized Coefficients
standardize_parameters(model_pref_level_financially_secure_successful_ambitious_robustcheck, method = "basic", ci = 0.997)## # A tibble: 3 × 5
## Parameter Std_Coefficient CI CI_low CI_high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0 0.997 0 0
## 2 political_orientation 0.110 0.997 0.0799 0.141
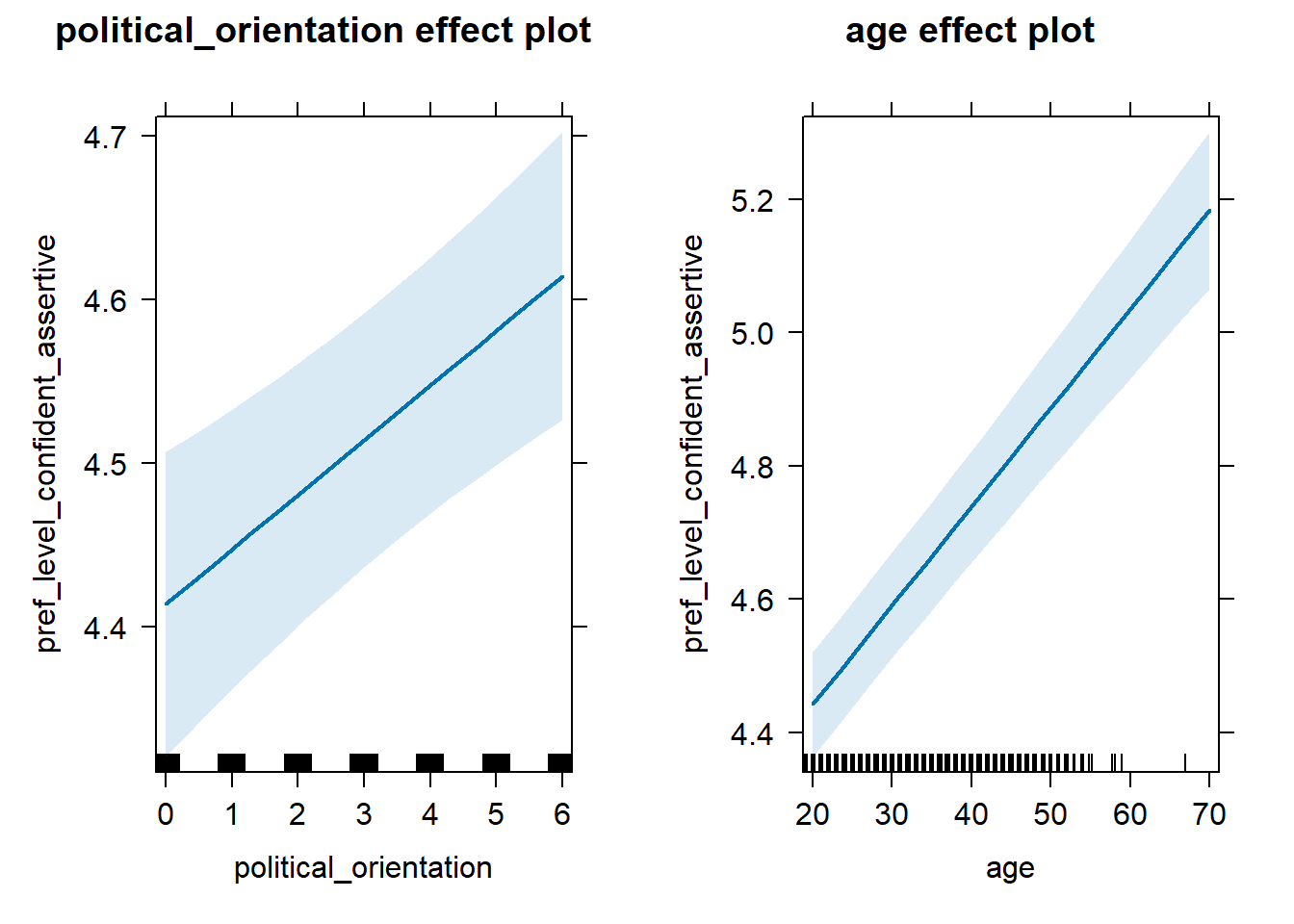
## 3 age 0.0724 0.997 0.0469 0.0978H2b Preference for the Level of Confidence-Assertiveness
H2b There is a positive linear link between right-wing political
orientation and women’s preferences for the level of confidence and
assertiveness.
Outcome: Level ratings for partner’s confidence-assertiveness.
Predictors: Political Orientation & Age. Random intercept and random
slope for country.
Models
Summary
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula: pref_level_confident_assertive ~ political_orientation + age +
## (1 + political_orientation | country)
## Data: data_included_documented
## Control: lmerControl(optimizer = "bobyqa")
##
## REML criterion at convergence: 28704.2
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -5.8511 -0.6602 -0.0056 0.6702 3.1600
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## country (Intercept) 0.1559960 0.39496
## political_orientation 0.0008448 0.02906 -0.54
## Residual 0.5512740 0.74248
## Number of obs: 12694, groups: country, 144
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 4.062e+00 5.346e-02 1.634e+02 75.99 < 2e-16 ***
## political_orientation 3.342e-02 7.958e-03 2.318e+01 4.20 0.000338 ***
## age 1.481e-02 9.729e-04 1.265e+04 15.22 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) pltcl_
## pltcl_rnttn -0.510
## age -0.456 0.036## Computing profile confidence intervals ...## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep## Warning in FUN(X[[i]], ...): non-monotonic profile for .sig02## Warning in confint.thpr(pp, level = level, zeta = zeta): bad spline fit for
## .sig02: falling back to linear interpolation## Warning in regularize.values(x, y, ties, missing(ties), na.rm = na.rm):
## Reduktion auf einmalige 'x' Werte## 0.15 % 99.85 %
## .sig01 0.293536620 0.53171540
## .sig02 -0.967408639 0.18411111
## .sig03 0.006596638 0.06129999
## .sigma 0.728761947 0.75657040
## (Intercept) 3.902864943 4.22349455
## political_orientation 0.007092236 0.05784956
## age 0.011922193 0.01769817Standardized Coefficients
standardize_parameters(model_pref_level_confident_assertive_robustcheck, method = "basic", ci = 0.997)## # A tibble: 3 × 5
## Parameter Std_Coefficient CI CI_low CI_high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0 0.997 0 0
## 2 political_orientation 0.0540 0.997 0.0158 0.0922
## 3 age 0.121 0.997 0.0976 0.145H2c Preference for the Level of Education-Intelligence
H2c There is no link between right-wing political orientation and
women’s preferences for the level of education and intelligence.
Outcome: Level ratings for partner’s education-intelligence. Predictors:
Political Orientation & Age. Random intercept and random slope for
country.
Models
Summary
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula: pref_level_intelligence_educated ~ political_orientation + age +
## (1 + political_orientation | country)
## Data: data_included_documented
## Control: lmerControl(optimizer = "bobyqa")
##
## REML criterion at convergence: 30053.3
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -5.2127 -0.6821 0.0344 0.6903 2.1844
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## country (Intercept) 0.0652437 0.25543
## political_orientation 0.0009699 0.03114 -0.24
## Residual 0.6129668 0.78292
## Number of obs: 12720, groups: country, 141
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 4.569e+00 4.586e-02 1.482e+02 99.625 <2e-16 ***
## political_orientation 2.328e-02 8.625e-03 2.888e+01 2.699 0.0115 *
## age 9.386e-03 1.020e-03 1.270e+04 9.198 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) pltcl_
## pltcl_rnttn -0.484
## age -0.554 0.030## Computing profile confidence intervals ...## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep## Warning in FUN(X[[i]], ...): non-monotonic profile for .sig02## Warning in confint.thpr(pp, level = level, zeta = zeta): bad spline fit for
## .sig02: falling back to linear interpolation## Warning in regularize.values(x, y, ties, missing(ties), na.rm = na.rm):
## Reduktion auf einmalige 'x' Werte## 0.15 % 99.85 %
## .sig01 0.171745467 0.37101593
## .sig02 -0.802102795 0.57378389
## .sig03 0.009368868 0.06422238
## .sigma 0.768481172 0.79774218
## (Intercept) 4.431590141 4.71156031
## political_orientation -0.003998975 0.05323338
## age 0.006355401 0.01241385Standardized Coefficients
standardize_parameters(model_pref_level_intelligence_educated_robustcheck, method = "basic", ci = 0.997)## # A tibble: 3 × 5
## Parameter Std_Coefficient CI CI_low CI_high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0 0.997 0 0
## 2 political_orientation 0.0386 0.997 -0.00384 0.0810
## 3 age 0.0791 0.997 0.0536 0.105Plot
lmer(pref_level_intelligence_educated ~ political_orientation + age + (1 + political_orientation|country),
data = data_included_documented) %>%
allEffects() %>%
plot()## Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
## Model failed to converge with max|grad| = 0.0107601 (tol = 0.002, component 1)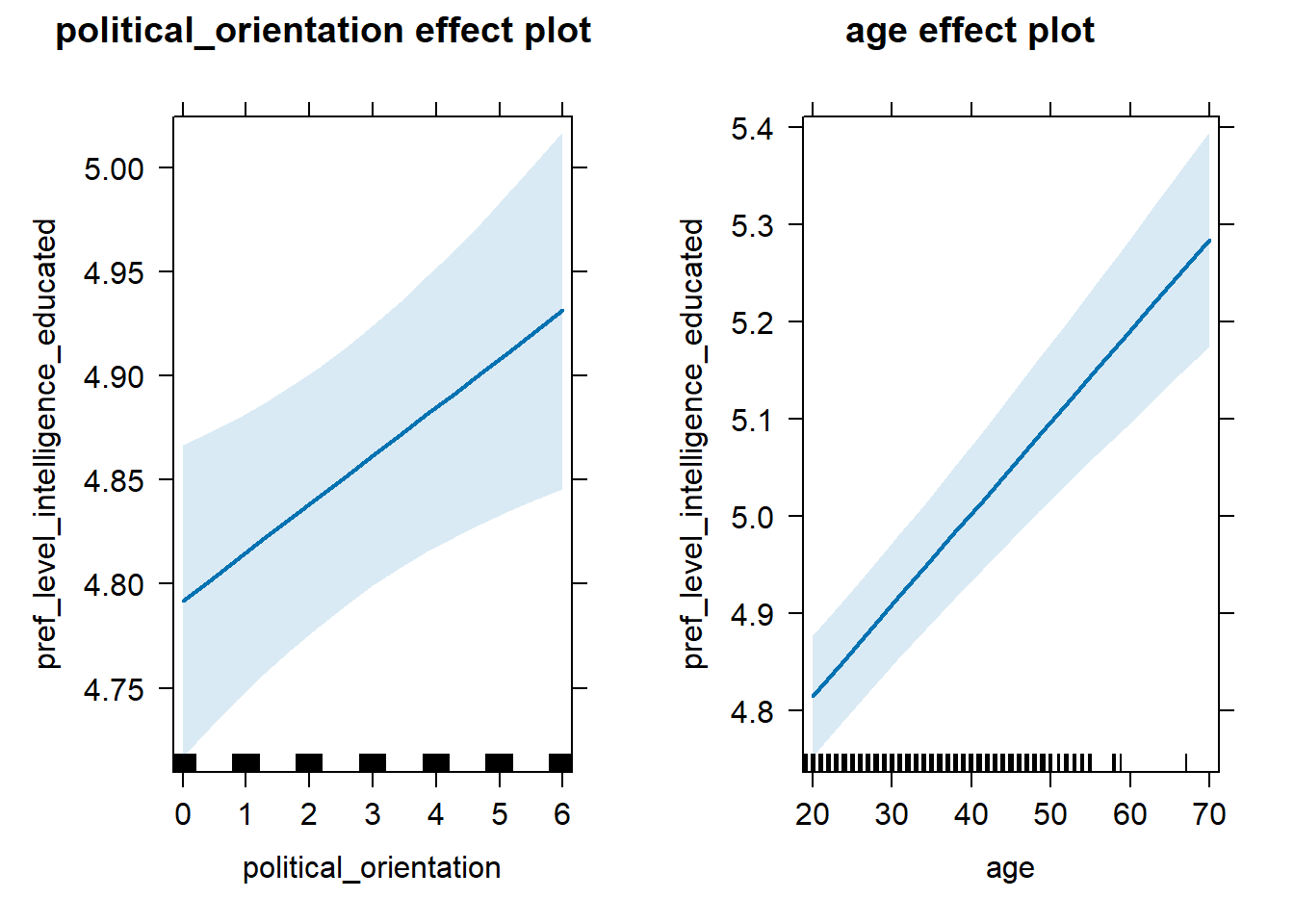
H2d Preference for the Level of Kindness-Supportiveness
H2d There is no link between right-wing political orientation and
women’s preferences for the level of kindness and supportiveness.
Outcome: Level ratings for partner’s kindness-supportiveness.
Predictors: Political Orientation & Age. Random intercept and random
slope for country.
Models
Summary
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula: pref_level_kind_supportive ~ political_orientation + age + (1 +
## political_orientation | country)
## Data: data_included_documented
## Control: lmerControl(optimizer = "bobyqa")
##
## REML criterion at convergence: 26526.1
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -4.9140 -0.6667 0.0855 0.8305 1.8003
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## country (Intercept) 1.615e-02 0.127072
## political_orientation 7.437e-05 0.008624 0.62
## Residual 4.663e-01 0.682850
## Number of obs: 12726, groups: country, 143
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 5.040e+00 3.163e-02 1.631e+02 159.327 < 2e-16 ***
## political_orientation 1.267e-02 4.891e-03 1.475e+01 2.590 0.0207 *
## age 4.959e-03 8.925e-04 1.266e+04 5.556 2.81e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) pltcl_
## pltcl_rnttn -0.323
## age -0.700 0.049## Computing profile confidence intervals ...## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep## Warning in FUN(X[[i]], ...): non-monotonic profile for .sig01## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
## identical or NA .zeta values: using minstep## Warning in FUN(X[[i]], ...): non-monotonic profile for .sig02## Warning in confint.thpr(pp, level = level, zeta = zeta): bad spline fit for
## .sig01: falling back to linear interpolation## Warning in confint.thpr(pp, level = level, zeta = zeta): bad spline fit for
## .sig02: falling back to linear interpolation## Warning in regularize.values(x, y, ties, missing(ties), na.rm = na.rm):
## Reduktion auf einmalige 'x' Werte## 0.15 % 99.85 %
## .sig01 0.101705922 0.204797206
## .sig02 -1.000000000 1.000000000
## .sig03 0.000000000 0.033780054
## .sigma 0.670273117 0.695760649
## (Intercept) 4.946227706 5.137257935
## political_orientation -0.003624704 0.029108389
## age 0.002304326 0.007608436Standardized Coefficients
## # A tibble: 3 × 5
## Parameter Std_Coefficient CI CI_low CI_high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0 0.997 0 0
## 2 political_orientation 0.0246 0.997 -0.00360 0.0529
## 3 age 0.0487 0.997 0.0227 0.0748Plot
lmer(pref_level_kind_supportive ~ political_orientation + age + (1 + political_orientation|country),
data = data_included_documented) %>%
allEffects() %>%
plot()## Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
## Model failed to converge with max|grad| = 0.00588346 (tol = 0.002, component 1)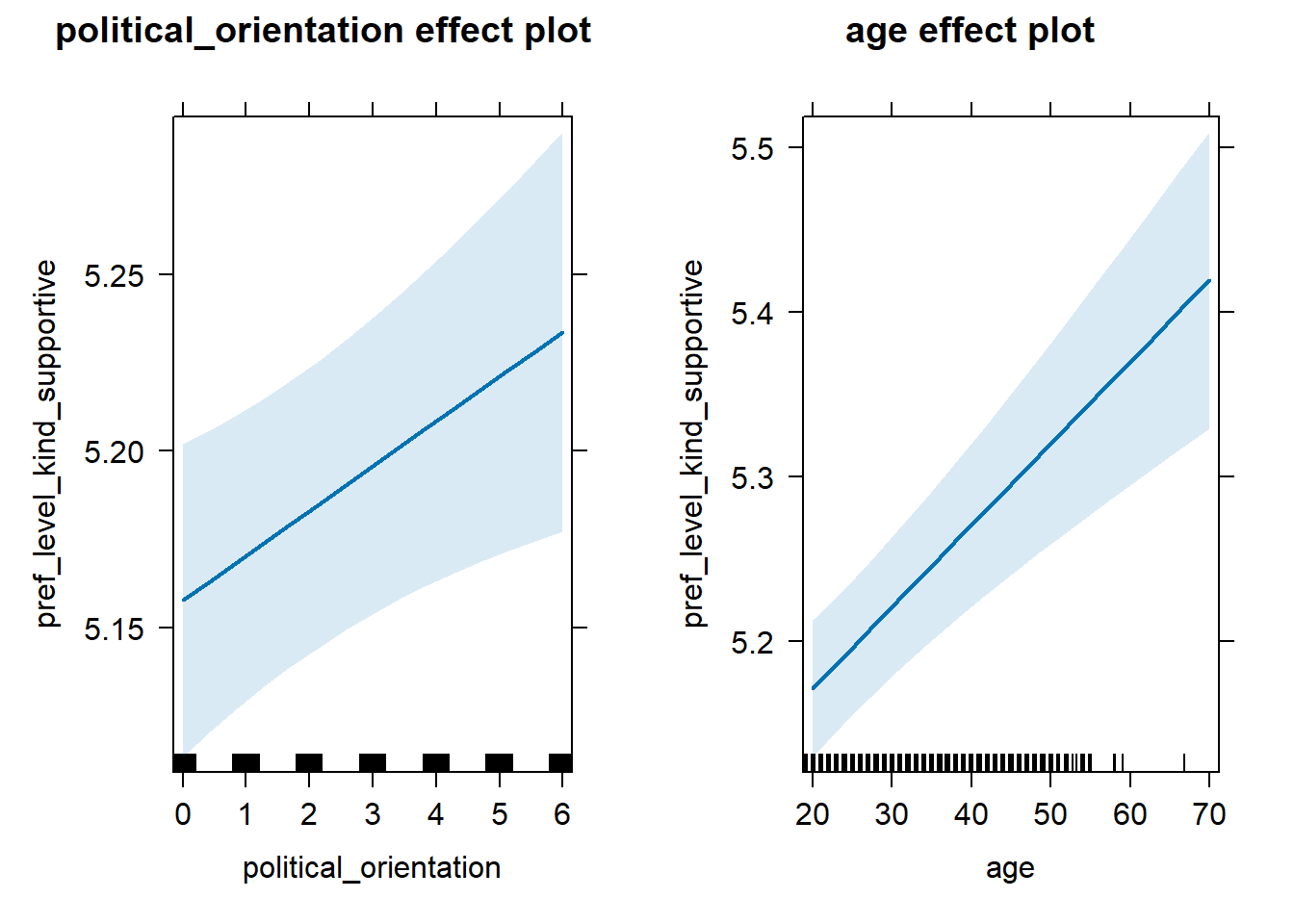
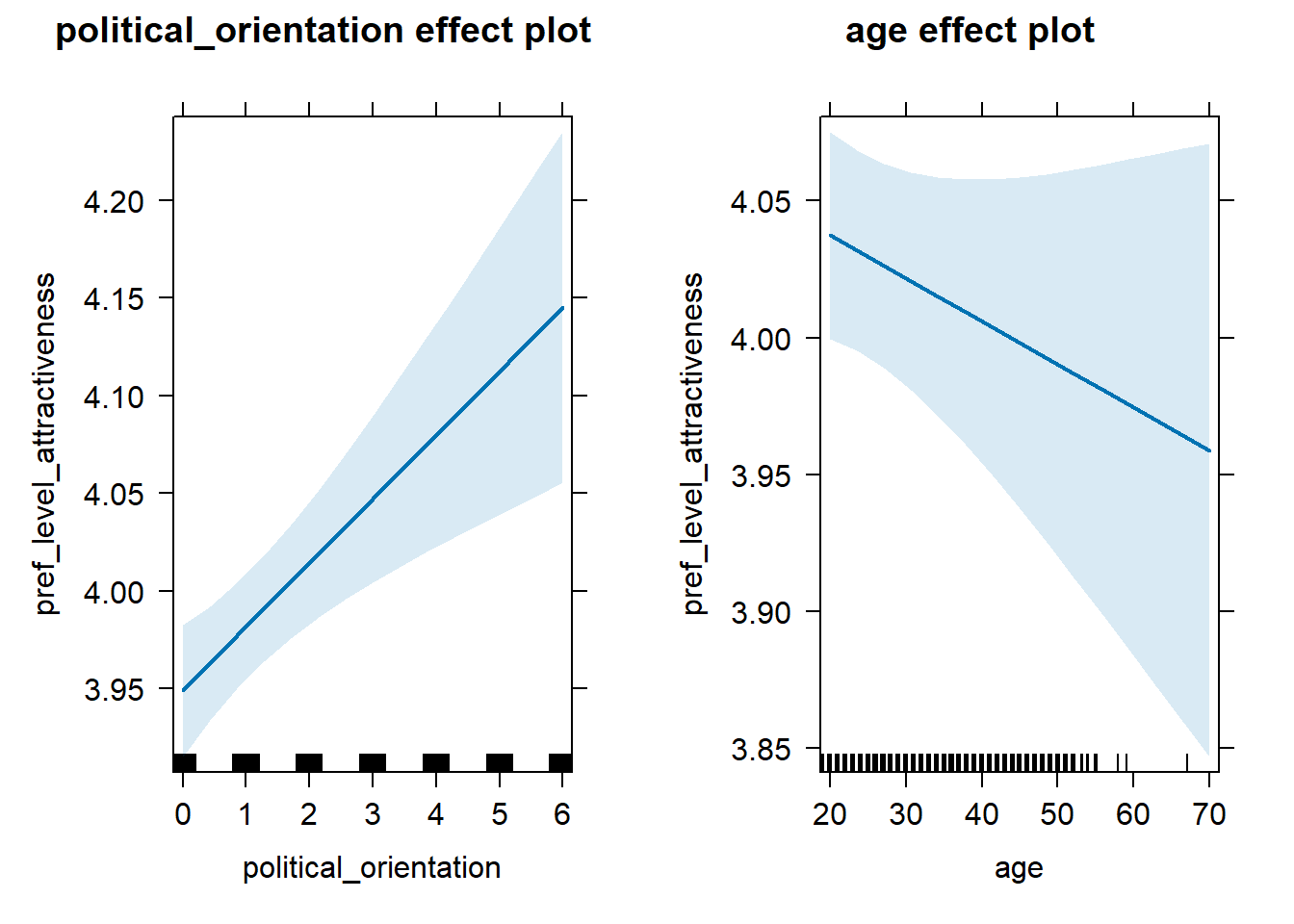
H2e Preference for the Level of Attractiveness
H2e There is no link between right-wing political orientation and
women’s preferences for the level of attractiveness.
Outcome: Level ratings for partner’s attractiveness. Predictors:
Political Orientation & Age. Random intercept and random slope for
country.
Models
Summary
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula: pref_level_attractiveness ~ political_orientation + age + (1 +
## political_orientation | country)
## Data: data_included_documented
## Control: lmerControl(optimizer = "bobyqa")
##
## REML criterion at convergence: 32911.1
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -4.6614 -0.6495 -0.0217 0.6475 2.5267
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## country (Intercept) 0.0109929 0.10485
## political_orientation 0.0002698 0.01643 0.27
## Residual 0.8049943 0.89721
## Number of obs: 12523, groups: country, 142
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 3.966e+00 3.864e-02 1.692e+02 102.616 < 2e-16 ***
## political_orientation 3.892e-02 7.120e-03 1.297e+01 5.466 0.000109 ***
## age -1.523e-03 1.174e-03 1.247e+04 -1.298 0.194326
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) pltcl_
## pltcl_rnttn -0.402
## age -0.751 0.041## Computing profile confidence intervals ...## 0.15 % 99.85 %
## .sig01 0.053277639 0.188794386
## .sig02 -0.822662479 1.000000000
## .sig03 0.000000000 0.058453744
## .sigma 0.880571880 0.914311109
## (Intercept) 3.850144942 4.081574661
## political_orientation 0.013284208 0.061250527
## age -0.005005251 0.001963986Standardized Coefficients
## # A tibble: 3 × 5
## Parameter Std_Coefficient CI CI_low CI_high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0 0.997 0 0
## 2 political_orientation 0.0579 0.997 0.0264 0.0893
## 3 age -0.0116 0.997 -0.0381 0.0149Ideal Age and Height
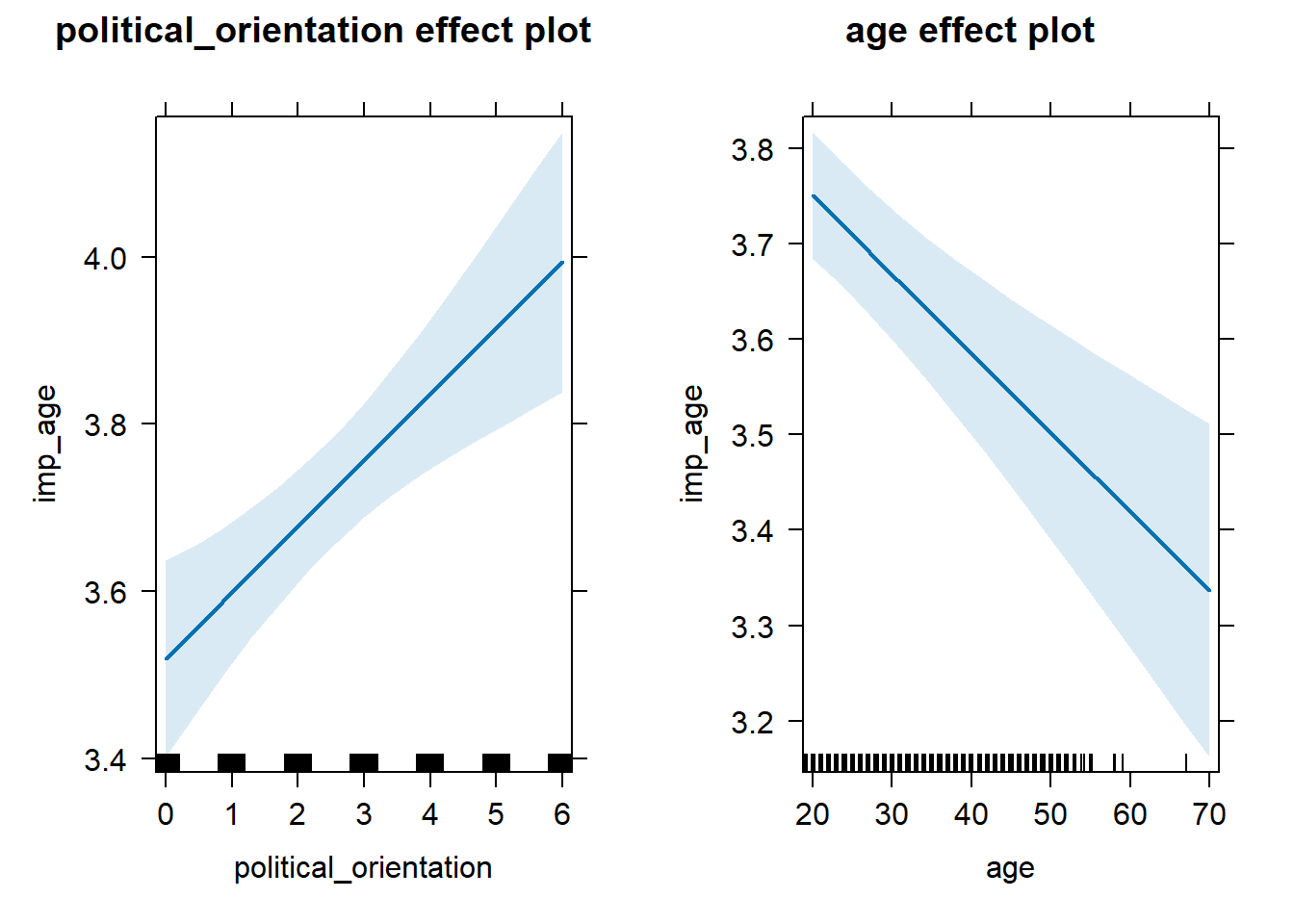
H3a(1) Importance Ratings for Partner’s Age
H3a(1) There is a positive linear link between right-wing political orientation and women’s importance ratings for partner’s age. Outcome: Importance ratings for partner’s age. Predictors: Political Orientation & Age. Random intercept and random slope for country.
Models
Summary
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula: imp_age ~ political_orientation + age + (1 + political_orientation |
## country)
## Data: data_included_documented
## Control: lmerControl(optimizer = "bobyqa")
##
## REML criterion at convergence: 46120.5
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -3.0860 -0.5065 0.1705 0.6534 2.1961
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## country (Intercept) 0.085465 0.29234
## political_orientation 0.009757 0.09878 -0.77
## Residual 1.956241 1.39866
## Number of obs: 13109, groups: country, 143
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 3.716e+00 7.452e-02 5.222e+01 49.859 < 2e-16 ***
## political_orientation 7.908e-02 2.032e-02 1.539e+01 3.892 0.00138 **
## age -8.266e-03 1.796e-03 1.307e+04 -4.603 4.21e-06 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) pltcl_
## pltcl_rnttn -0.683
## age -0.597 0.024## Computing profile confidence intervals ...## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep## Warning in FUN(X[[i]], ...): non-monotonic profile for .sig01## Warning in confint.thpr(pp, level = level, zeta = zeta): bad spline fit for
## .sig01: falling back to linear interpolation## 0.15 % 99.85 %
## .sig01 0.110868701 0.560820496
## .sig02 -0.961434270 0.617046419
## .sig03 0.007766235 0.203196740
## .sigma 1.373282066 1.424910245
## (Intercept) 3.486463009 3.959721045
## political_orientation 0.011026257 0.147576380
## age -0.013606154 -0.002940114Standardized Coefficients
## # A tibble: 3 × 5
## Parameter Std_Coefficient CI CI_low CI_high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0 0.997 0 0
## 2 political_orientation 0.0759 0.997 0.0180 0.134
## 3 age -0.0402 0.997 -0.0661 -0.0143H3a(2) Level Ratings for Partner’s Age
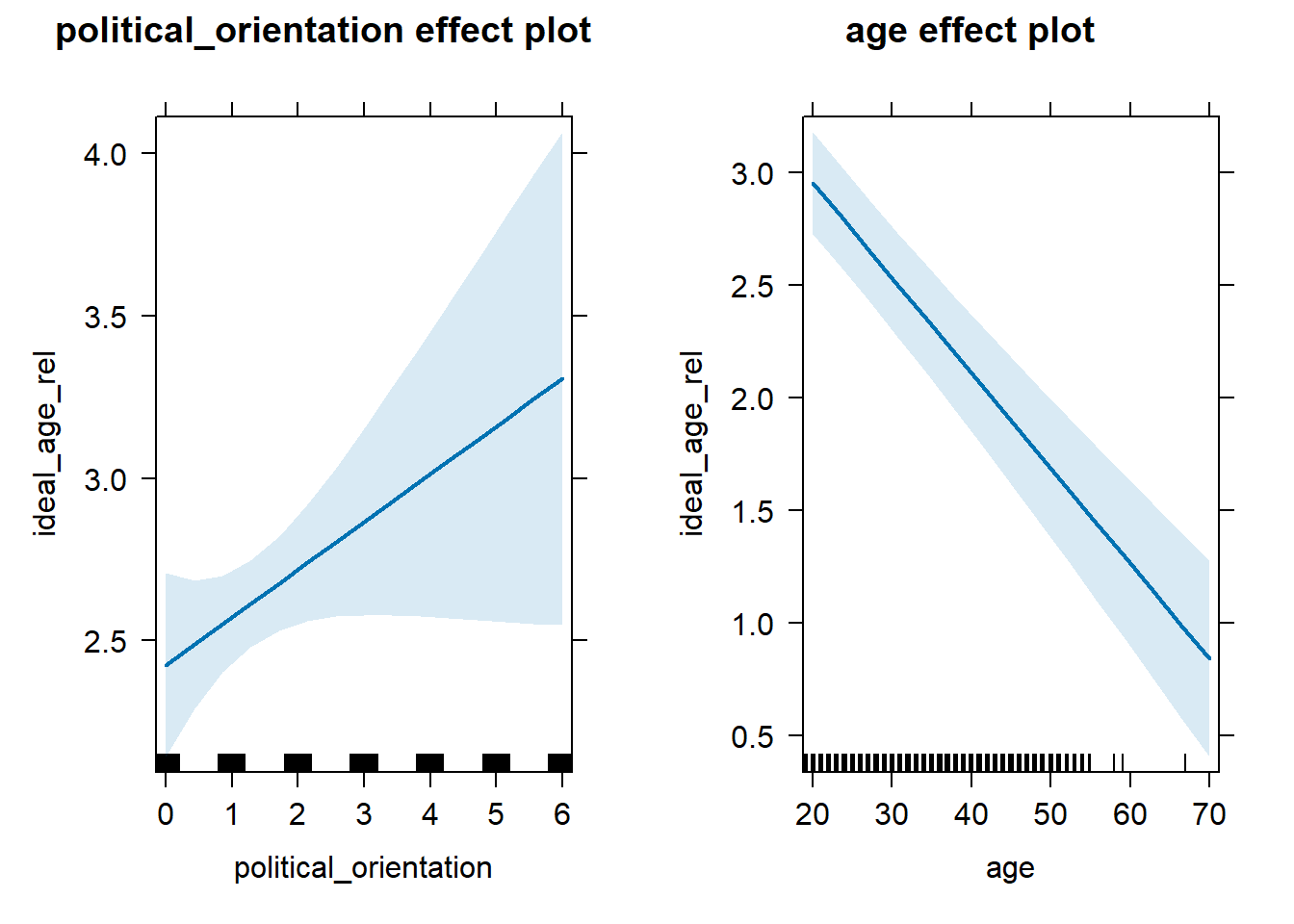
H3a(2) There is a positive linear link between right-wing political orientation and the relative age discrepancy between ideal partner’s age and women’s age. (discrepancy calculated as ideal partner’s age – women’s age) Outcome: Discrepancy between level ratings for ideal partner’s age and women’s age (ideal_age_rel) Predictors: Political Orientation & Age. Random intercept and random slope for country.
Models
Summary
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula:
## ideal_age_rel ~ political_orientation + age + (1 + political_orientation |
## country)
## Data: data_included_documented
## Control: lmerControl(optimizer = "bobyqa")
##
## REML criterion at convergence: 61595.1
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -11.4254 -0.4968 -0.1225 0.4167 25.7466
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## country (Intercept) 0.8110 0.9006
## political_orientation 0.4583 0.6770 -0.94
## Residual 9.5160 3.0848
## Number of obs: 12057, groups: country, 140
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 3.425e+00 1.762e-01 3.244e+01 19.443 <2e-16 ***
## political_orientation 1.469e-01 8.420e-02 3.640e+01 1.745 0.0894 .
## age -4.221e-02 4.158e-03 1.199e+04 -10.152 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) pltcl_
## pltcl_rnttn -0.711
## age -0.576 0.006## Computing profile confidence intervals ...## 0.15 % 99.85 %
## .sig01 0.23944723 1.86330535
## .sig02 -0.99282843 0.01939224
## .sig03 0.39869760 1.01469699
## .sigma 3.02631366 3.14499654
## (Intercept) 2.82919847 3.98186213
## political_orientation -0.11315423 0.41406585
## age -0.05456051 -0.02987641Standardized Coefficients
## # A tibble: 3 × 5
## Parameter Std_Coefficient CI CI_low CI_high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0 0.997 0 0
## 2 political_orientation 0.0636 0.997 -0.0446 0.172
## 3 age -0.0921 0.997 -0.119 -0.0652H3b(1) Importance Ratings for Partner’s Height
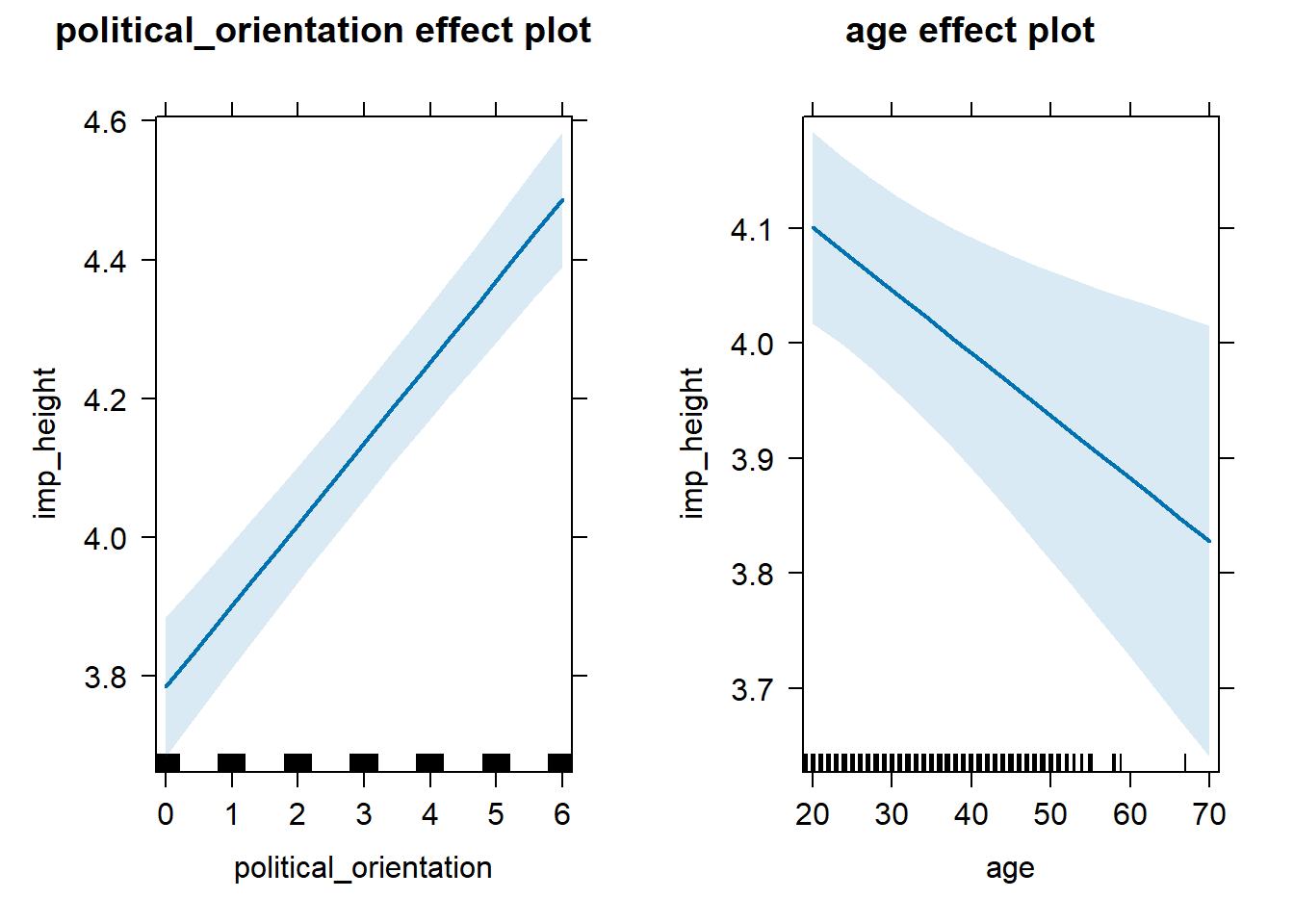
H3b(1) There is a positive linear link between right-wing political orientation and women’s importance ratings for partner’s height. Outcome: Importance ratings for partner’s height. Predictors: Political Orientation & Age. Random intercept and random slope for country.
Models
model_imp_height_robustcheck <- lmer(imp_height ~ political_orientation + age + (1+political_orientation|country), data = data_included_documented, control =lmerControl(optimizer = "bobyqa"))## boundary (singular) fit: see help('isSingular')Summary
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula:
## imp_height ~ political_orientation + age + (1 + political_orientation |
## country)
## Data: data_included_documented
## Control: lmerControl(optimizer = "bobyqa")
##
## REML criterion at convergence: 46906
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -3.2839 -0.5568 0.0992 0.7234 2.0290
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## country (Intercept) 8.664e-02 0.294348
## political_orientation 7.693e-05 0.008771 -1.00
## Residual 2.130e+00 1.459394
## Number of obs: 13022, groups: country, 143
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 3.913e+00 6.934e-02 1.647e+02 56.43 < 2e-16 ***
## political_orientation 1.171e-01 9.667e-03 6.703e+02 12.12 < 2e-16 ***
## age -5.444e-03 1.877e-03 1.302e+04 -2.90 0.00374 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) pltcl_
## pltcl_rnttn -0.476
## age -0.674 0.057
## optimizer (bobyqa) convergence code: 0 (OK)
## boundary (singular) fit: see help('isSingular')## Computing profile confidence intervals ...## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep## Warning in FUN(X[[i]], ...): non-monotonic profile for .sig01## Warning in FUN(X[[i]], ...): non-monotonic profile for .sig02## Warning in confint.thpr(pp, level = level, zeta = zeta): bad spline fit for
## .sig01: falling back to linear interpolation## Warning in confint.thpr(pp, level = level, zeta = zeta): bad spline fit for
## .sig02: falling back to linear interpolation## 0.15 % 99.85 %
## .sig01 0.18244127 0.3923682822
## .sig02 -1.00000000 1.0000000000
## .sig03 0.00000000 0.0741722096
## .sigma 1.43364813 1.4865977521
## (Intercept) 3.70585092 4.1346728259
## political_orientation 0.08195292 0.1514853491
## age -0.01102432 0.0001264944Standardized Coefficients
## # A tibble: 3 × 5
## Parameter Std_Coefficient CI CI_low CI_high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0 0.997 0 0
## 2 political_orientation 0.107 0.997 0.0808 0.133
## 3 age -0.0252 0.997 -0.0510 0.000597H3b(2) Level Ratings for Partner’s Height
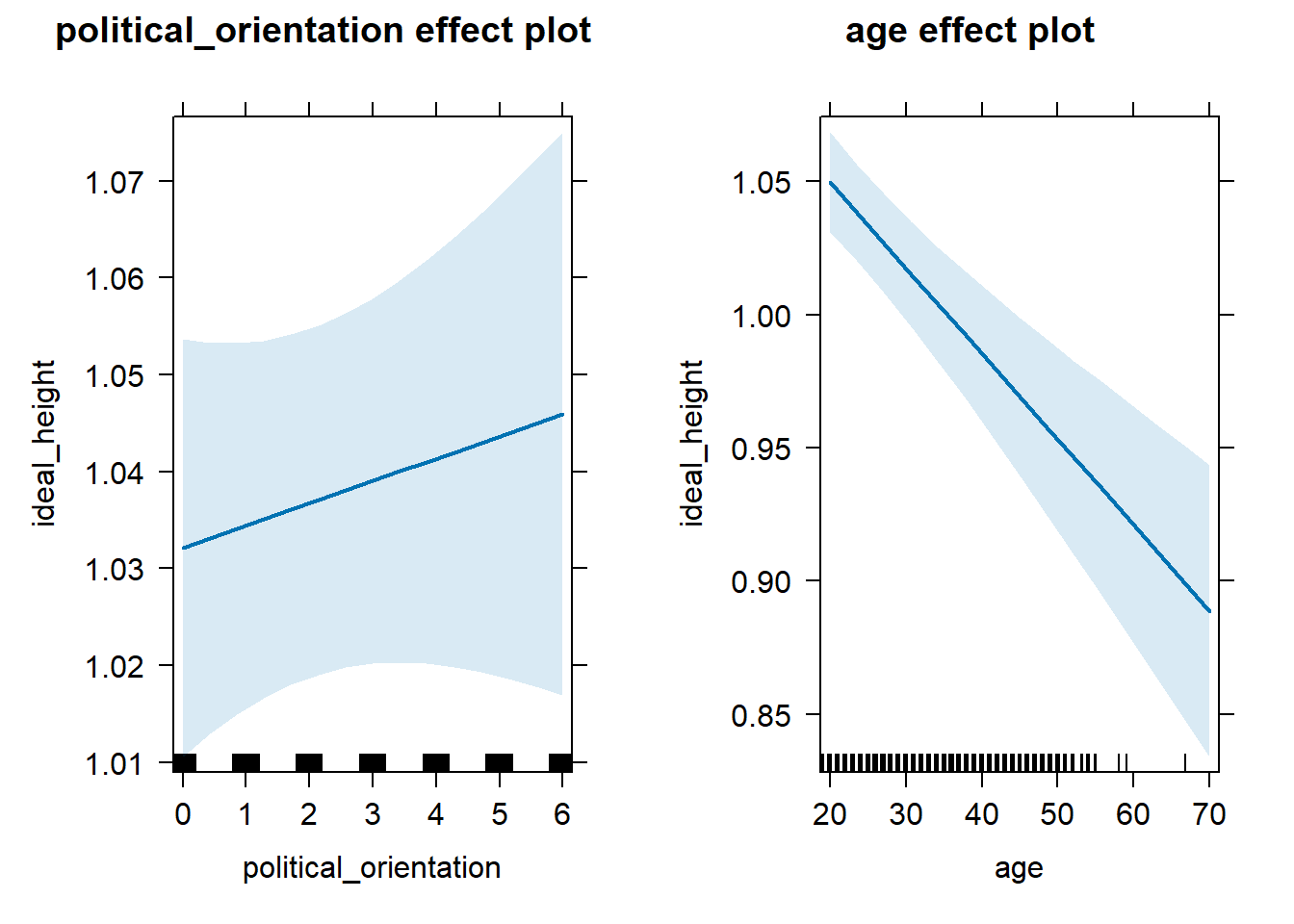
H3b(2) There is a positive linear link between right-wing political orientation and ideal partner’s height. Outcome: Level ratings for ideal partner’s height. Predictors: Political Orientation & Age. Random intercept and random slope for country.
Models
model_ideal_height_robustcheck <- lmer(ideal_height ~ political_orientation + age + (1+political_orientation|country), data = data_included_documented, control = lmerControl(optimizer = "bobyqa"))## boundary (singular) fit: see help('isSingular')Summary
## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula:
## ideal_height ~ political_orientation + age + (1 + political_orientation |
## country)
## Data: data_included_documented
## Control: lmerControl(optimizer = "bobyqa")
##
## REML criterion at convergence: 14661.3
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -7.1159 -0.1374 -0.0759 0.0026 2.5926
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## country (Intercept) 1.685e-03 0.041047
## political_orientation 1.134e-05 0.003368 1.00
## Residual 1.877e-01 0.433192
## Number of obs: 12520, groups: country, 142
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 1.108e+00 1.783e-02 2.196e+02 62.172 < 2e-16 ***
## political_orientation 2.312e-03 2.945e-03 3.601e+02 0.785 0.433
## age -3.220e-03 5.715e-04 1.246e+04 -5.634 1.8e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) pltcl_
## pltcl_rnttn -0.371
## age -0.788 0.044
## optimizer (bobyqa) convergence code: 0 (OK)
## boundary (singular) fit: see help('isSingular')## Computing profile confidence intervals ...## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep
## Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
## profile: using minstep## Warning in FUN(X[[i]], ...): non-monotonic profile for .sig01## Warning in FUN(X[[i]], ...): non-monotonic profile for .sig02## Warning in confint.thpr(pp, level = level, zeta = zeta): bad spline fit for
## .sig01: falling back to linear interpolation## Warning in confint.thpr(pp, level = level, zeta = zeta): bad spline fit for
## .sig02: falling back to linear interpolation## 0.15 % 99.85 %
## .sig01 0.015219409 0.085389183
## .sig02 -1.000000000 1.000000000
## .sig03 0.000000000 0.015443341
## .sigma 0.475998562 0.445652997
## (Intercept) 1.055198315 1.164038633
## political_orientation -0.008530965 0.011898733
## age -0.004914860 -0.001521768Standardized Coefficients
## # A tibble: 3 × 5
## Parameter Std_Coefficient CI CI_low CI_high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0 0.997 0 0
## 2 political_orientation 0.00720 0.997 -0.0200 0.0344
## 3 age -0.0506 0.997 -0.0772 -0.0239