3_sample_analyses
Laura Botzet & Ruben Arslan
Sample Analyses
Helper
source("0_helpers.R")##
## Attaching package: 'formr'## The following object is masked from 'package:rmarkdown':
##
## word_document##
## Attaching package: 'lubridate'## The following object is masked from 'package:base':
##
## date## Loading required package: carData## lattice theme set by effectsTheme()
## See ?effectsTheme for details.##
## Attaching package: 'data.table'## The following objects are masked from 'package:lubridate':
##
## hour, isoweek, mday, minute, month, quarter, second, wday, week, yday, year## The following objects are masked from 'package:formr':
##
## first, last## Loading required package: Matrix##
## Attaching package: 'lmerTest'## The following object is masked from 'package:lme4':
##
## lmer## The following object is masked from 'package:stats':
##
## step##
## Attaching package: 'psych'## The following objects are masked from 'package:ggplot2':
##
## %+%, alpha## This is lavaan 0.6-3## lavaan is BETA software! Please report any bugs.##
## Attaching package: 'lavaan'## The following object is masked from 'package:psych':
##
## cor2cov## Loading required package: lattice## Loading required package: survival## Loading required package: Formula##
## Attaching package: 'Hmisc'## The following object is masked from 'package:psych':
##
## describe## The following objects are masked from 'package:base':
##
## format.pval, units##
## Attaching package: 'tidyr'## The following object is masked from 'package:Matrix':
##
## expand##
## Attaching package: 'dplyr'## The following objects are masked from 'package:Hmisc':
##
## src, summarize## The following objects are masked from 'package:data.table':
##
## between, first, last## The following objects are masked from 'package:lubridate':
##
## intersect, setdiff, union## The following objects are masked from 'package:formr':
##
## first, last## The following objects are masked from 'package:stats':
##
## filter, lag## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union##
## Attaching package: 'codebook'## The following object is masked from 'package:psych':
##
## bfi## The following objects are masked from 'package:formr':
##
## aggregate_and_document_scale, asis_knit_child, expired, paste.knit_asis##
## Attaching package: 'effsize'## The following object is masked from 'package:psych':
##
## cohen.dLoad data
### Import all data with known birthorder
birthorder = readRDS("data/alldata_birthorder.rds")Overview Raw data
# codebook(birthorder)Data wrangling
## we have to exclude people in the control group who are part of the birthorder group
birthorder = birthorder %>%
# mark people who have missing birthorder data
mutate(check_birthorder = ifelse(!is.na(birthorder_genes), 1, 0),
# mark people who have missing outcomes
check_outcome = ifelse(!is.na(raven_2015_old), 1,
ifelse(!is.na(math_2015_old), 1,
ifelse(!is.na(raven_2015_young), 1,
ifelse(!is.na(math_2015_young), 1,
ifelse(!is.na(raven_2007_old), 1,
ifelse(!is.na(math_2007_old), 1,
ifelse(!is.na(raven_2007_young), 1,
ifelse(!is.na(math_2007_young), 1,
ifelse(!is.na(adaptive_numbering), 1,
ifelse(!is.na(words_remembered_avg), 1,
ifelse(!is.na(count_backwards), 1,
ifelse(!is.na(big5_ext), 1,
ifelse(!is.na(riskA), 1,
ifelse(!is.na(riskB), 1,
ifelse(!is.na(years_of_education), 1,
ifelse(!is.na(Elementary_missed), 1,
ifelse(!is.na(Elementary_worked), 1,
ifelse(!is.na(attended_school), 1,
ifelse(!is.na(wage_last_month_log), 1,
ifelse(!is.na(wage_last_year_log), 1,
ifelse(!is.na(Self_employed), 1,
ifelse(!is.na(Category), 1,
ifelse(!is.na(Sector), 1,
ifelse(!is.na(ever_smoked), 1,
ifelse(!is.na(still_smoking), 1,
0))))))))))))))))))))))))),
group_birthorder = ifelse(check_birthorder == 0, "control", "test"),
group_outcome = ifelse(check_outcome == 0, "control", "test"))Birth order and sibship size
Genes birthorder
descriptives = birthorder %>%
group_by(group_outcome) %>%
summarise(individuals = n(),
mothers = length(unique(mother_pidlink)),
sibship_size_mean = mean(sibling_count_genes, na.rm = T),
sibship_size_confidence_low = mean(sibling_count_genes, na.rm = T) -
(qt(.975, n()-1)*sd(sibling_count_genes, na.rm = T)/sqrt(n())),
sibship_size_confidence_high = mean(sibling_count_genes, na.rm = T) +
(qt(.975, n()-1)*sd(sibling_count_genes, na.rm = T)/sqrt(n())),
sibship_size_min = min(sibling_count_genes, na.rm = TRUE),
sibship_size_max = max(sibling_count_genes, na.rm = TRUE),
birthorder_mean = mean(birthorder_genes, na.rm = T),
birthorder_size_confidence_low = mean(birthorder_genes, na.rm = T) -
(qt(.975, n()-1)*sd(birthorder_genes, na.rm = T)/sqrt(n())),
birthorder_size_confidence_high = mean(birthorder_genes, na.rm = T) +
(qt(.975, n()-1)*sd(birthorder_genes, na.rm = T)/sqrt(n())),
birthorder_size_min = min(birthorder_genes, na.rm = TRUE),
birthorder_size_max = max(birthorder_genes, na.rm = TRUE),
number_siblings_mean = mean(sibling_count_genes, na.rm =T),
number_siblings_confidence_low = mean(sibling_count_genes, na.rm = T) -
(qt(.975, n()-1)*sd(sibling_count_genes, na.rm = T)/sqrt(n())),
number_siblings_confidence_high = mean(sibling_count_genes, na.rm = T) +
(qt(.975, n()-1)*sd(sibling_count_genes, na.rm = T)/sqrt(n())))
descriptives| group_outcome | individuals | mothers | sibship_size_mean | sibship_size_confidence_low | sibship_size_confidence_high | sibship_size_min | sibship_size_max | birthorder_mean | birthorder_size_confidence_low | birthorder_size_confidence_high | birthorder_size_min | birthorder_size_max | number_siblings_mean | number_siblings_confidence_low | number_siblings_confidence_high |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| control | 55765 | 16301 | 4.422 | 4.396 | 4.448 | 1 | 22 | 2.689 | 2.672 | 2.707 | 1 | 22 | 4.422 | 4.396 | 4.448 |
| test | 45476 | 13187 | 3.702 | 3.68 | 3.724 | 1 | 22 | 2.374 | 2.357 | 2.391 | 1 | 21 | 3.702 | 3.68 | 3.724 |
birthorder %>%
t.test(sibling_count_genes ~ group_outcome, data = ., var.equal = T)| Test statistic | df | P value | Alternative hypothesis | mean in group control | mean in group test |
|---|---|---|---|---|---|
| 24.32 | 42680 | 9.738e-130 * * * | two.sided | 4.422 | 3.702 |
birthorder %>%
t.test(birthorder_genes ~ group_outcome, data = ., var.equal = T)| Test statistic | df | P value | Alternative hypothesis | mean in group control | mean in group test |
|---|---|---|---|---|---|
| 15.13 | 42680 | 1.458e-51 * * * | two.sided | 2.689 | 2.374 |
How many siblings with data do we retain in each family?
birthorder %>% filter(!is.na(birthorder_genes), group_outcome == "test") %>% group_by(mother_pidlink) %>% summarise(with_data = n(), all = mean(sibling_count_genes)) -> counts
birthorder %>% filter(!is.na(birthorder_genes), group_outcome == "control") %>% group_by(mother_pidlink) %>% summarise(with_data = n(), all = mean(sibling_count_genes)) -> counts1In our test sample families with an average size of 3.1623 siblings, we retain 1.7438.
In the original sample families with an average size of 2.896 siblings, we retain 2.2035.
ggplot(counts, aes(all, with_data)) + geom_jitter(alpha = 0.1) + geom_smooth() + scale_x_continuous(breaks=1:15) + scale_y_continuous(breaks=1:15)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'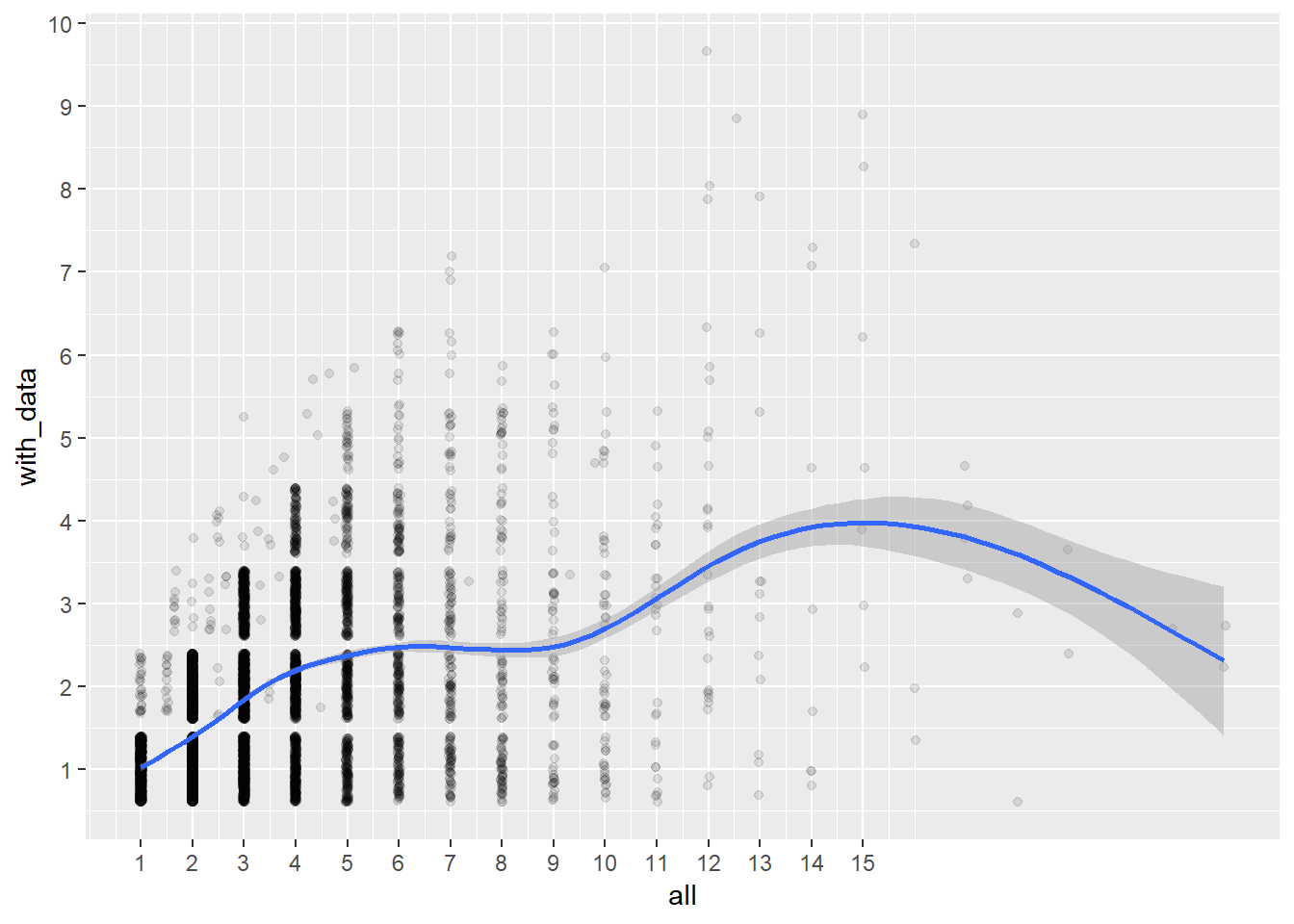
Maternal birthorder
descriptives = birthorder %>%
group_by(group_outcome) %>%
summarise(individuals = n(),
mothers = length(unique(mother_pidlink)),
sibship_size_mean = mean(sibling_count_uterus_alive, na.rm = T),
sibship_size_confidence_low = mean(sibling_count_uterus_alive, na.rm = T) -
(qt(.975, n()-1)*sd(sibling_count_uterus_alive, na.rm = T)/sqrt(n())),
sibship_size_confidence_high = mean(sibling_count_uterus_alive, na.rm = T) +
(qt(.975, n()-1)*sd(sibling_count_uterus_alive, na.rm = T)/sqrt(n())),
sibship_size_min = min(sibling_count_uterus_alive, na.rm = TRUE),
sibship_size_max = max(sibling_count_uterus_alive, na.rm = TRUE),
birthorder_mean = mean(birthorder_uterus_alive, na.rm = T),
birthorder_size_confidence_low = mean(birthorder_uterus_alive, na.rm = T) -
(qt(.975, n()-1)*sd(birthorder_uterus_alive, na.rm = T)/sqrt(n())),
birthorder_size_confidence_high = mean(birthorder_uterus_alive, na.rm = T) +
(qt(.975, n()-1)*sd(birthorder_uterus_alive, na.rm = T)/sqrt(n())),
birthorder_size_min = min(birthorder_uterus_alive, na.rm = TRUE),
birthorder_size_max = max(birthorder_uterus_alive, na.rm = TRUE),
number_siblings_mean = mean(sibling_count_uterus_alive, na.rm =T),
number_siblings_confidence_low = mean(sibling_count_uterus_alive, na.rm = T) -
(qt(.975, n()-1)*sd(sibling_count_uterus_alive, na.rm = T)/sqrt(n())),
number_siblings_confidence_high = mean(sibling_count_uterus_alive, na.rm = T) +
(qt(.975, n()-1)*sd(sibling_count_uterus_alive, na.rm = T)/sqrt(n())))
descriptives| group_outcome | individuals | mothers | sibship_size_mean | sibship_size_confidence_low | sibship_size_confidence_high | sibship_size_min | sibship_size_max | birthorder_mean | birthorder_size_confidence_low | birthorder_size_confidence_high | birthorder_size_min | birthorder_size_max | number_siblings_mean | number_siblings_confidence_low | number_siblings_confidence_high |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| control | 55765 | 16301 | 4.614 | 4.587 | 4.641 | 1 | 22 | 2.775 | 2.757 | 2.793 | 1 | 22 | 4.614 | 4.587 | 4.641 |
| test | 45476 | 13187 | 3.823 | 3.8 | 3.845 | 1 | 22 | 2.454 | 2.436 | 2.471 | 1 | 21 | 3.823 | 3.8 | 3.845 |
birthorder %>%
t.test(sibling_count_uterus_alive ~ group_outcome, data = ., var.equal = T)| Test statistic | df | P value | Alternative hypothesis | mean in group control | mean in group test |
|---|---|---|---|---|---|
| 26.37 | 43320 | 5.226e-152 * * * | two.sided | 4.614 | 3.823 |
birthorder %>%
t.test(birthorder_uterus_alive ~ group_outcome, data = ., var.equal = T)| Test statistic | df | P value | Alternative hypothesis | mean in group control | mean in group test |
|---|---|---|---|---|---|
| 15.08 | 43320 | 2.809e-51 * * * | two.sided | 2.775 | 2.454 |
How many siblings with data do we retain in each family?
birthorder %>% filter(!is.na(birthorder_uterus_alive), group_outcome == 1) %>% group_by(mother_pidlink) %>% summarise(with_data = n(), all = mean(sibling_count_uterus_alive)) -> counts
birthorder %>% filter(!is.na(birthorder_uterus_alive), group_outcome == 0) %>% group_by(mother_pidlink) %>% summarise(with_data = n(), all = mean(sibling_count_uterus_alive)) -> counts1In our test sample families with an average size of NaN siblings, we retain 0.
In the original sample families with an average size of NaN siblings, we retain 0.
ggplot(counts, aes(all, with_data)) + geom_jitter(alpha = 0.1) + geom_smooth() + scale_x_continuous(breaks=1:15) + scale_y_continuous(breaks=1:15)## `geom_smooth()` using method = 'loess' and formula 'y ~ x' ### Maternal pregnancy birthorder
### Maternal pregnancy birthorder
descriptives = birthorder %>%
group_by(group_outcome) %>%
summarise(individuals = n(),
mothers = length(unique(mother_pidlink)),
sibship_size_mean = mean(sibling_count_uterus_preg, na.rm = T),
sibship_size_confidence_low = mean(sibling_count_uterus_preg, na.rm = T) -
(qt(.975, n()-1)*sd(sibling_count_uterus_preg, na.rm = T)/sqrt(n())),
sibship_size_confidence_high = mean(sibling_count_uterus_preg, na.rm = T) +
(qt(.975, n()-1)*sd(sibling_count_uterus_preg, na.rm = T)/sqrt(n())),
sibship_size_min = min(sibling_count_uterus_preg, na.rm = TRUE),
sibship_size_max = max(sibling_count_uterus_preg, na.rm = TRUE),
birthorder_mean = mean(birthorder_uterus_preg, na.rm = T),
birthorder_size_confidence_low = mean(birthorder_uterus_preg, na.rm = T) -
(qt(.975, n()-1)*sd(birthorder_uterus_preg, na.rm = T)/sqrt(n())),
birthorder_size_confidence_high = mean(birthorder_uterus_preg, na.rm = T) +
(qt(.975, n()-1)*sd(birthorder_uterus_preg, na.rm = T)/sqrt(n())),
birthorder_size_min = min(birthorder_uterus_preg, na.rm = TRUE),
birthorder_size_max = max(birthorder_uterus_preg, na.rm = TRUE),
number_siblings_mean = mean(sibling_count_uterus_preg, na.rm =T),
number_siblings_confidence_low = mean(sibling_count_uterus_preg, na.rm = T) -
(qt(.975, n()-1)*sd(sibling_count_uterus_preg, na.rm = T)/sqrt(n())),
number_siblings_confidence_high = mean(sibling_count_uterus_preg, na.rm = T) +
(qt(.975, n()-1)*sd(sibling_count_uterus_preg, na.rm = T)/sqrt(n())))
descriptives| group_outcome | individuals | mothers | sibship_size_mean | sibship_size_confidence_low | sibship_size_confidence_high | sibship_size_min | sibship_size_max | birthorder_mean | birthorder_size_confidence_low | birthorder_size_confidence_high | birthorder_size_min | birthorder_size_max | number_siblings_mean | number_siblings_confidence_low | number_siblings_confidence_high |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| control | 55765 | 16301 | 5.184 | 5.154 | 5.214 | 1 | 26 | 3.072 | 3.052 | 3.093 | 1 | 26 | 5.184 | 5.154 | 5.214 |
| test | 45476 | 13187 | 4.279 | 4.253 | 4.305 | 1 | 26 | 2.663 | 2.644 | 2.683 | 1 | 25 | 4.279 | 4.253 | 4.305 |
birthorder %>%
t.test(sibling_count_uterus_preg ~ group_outcome, data = ., var.equal = T)| Test statistic | df | P value | Alternative hypothesis | mean in group control | mean in group test |
|---|---|---|---|---|---|
| 27.27 | 48544 | 1.473e-162 * * * | two.sided | 5.184 | 4.279 |
birthorder %>%
t.test(birthorder_uterus_preg ~ group_outcome, data = ., var.equal = T)| Test statistic | df | P value | Alternative hypothesis | mean in group control | mean in group test |
|---|---|---|---|---|---|
| 17.36 | 48544 | 2.76e-67 * * * | two.sided | 3.072 | 2.663 |
How many siblings with data do we retain in each family?
birthorder %>% filter(!is.na(birthorder_uterus_preg), group_outcome == 1) %>% group_by(mother_pidlink) %>% summarise(with_data = n(), all = mean(sibling_count_uterus_preg)) -> counts
birthorder %>% filter(!is.na(birthorder_uterus_preg), group_outcome == 0) %>% group_by(mother_pidlink) %>% summarise(with_data = n(), all = mean(sibling_count_uterus_preg)) -> counts1In our test sample families with an average size of NaN siblings, we retain 0.
In the original sample families with an average size of NaN siblings, we retain 0.
ggplot(counts, aes(all, with_data)) + geom_jitter(alpha = 0.1) + geom_smooth() + scale_x_continuous(breaks=1:15) + scale_y_continuous(breaks=1:15)## `geom_smooth()` using method = 'loess' and formula 'y ~ x'
Outcome measurements and covariates
Control group
## Descriptives
descriptives = birthorder %>%
group_by(group_birthorder) %>%
summarise(n = n(),
age_mean = mean(age, na.rm=TRUE),
age_confidence_low = mean(age, na.rm = T) -
(qt(.975, n()-1)*sd(age, na.rm = T)/sqrt(n())),
age_confidence_high = mean(age, na.rm = T) +
(qt(.975, n()-1)*sd(age, na.rm = T)/sqrt(n())),
age_min = min(age, na.rm = TRUE),
age_max = max(age, na.rm = TRUE),
gender = mean(male, na.rm=TRUE))
descriptives| group_birthorder | n | age_mean | age_confidence_low | age_confidence_high | age_min | age_max | gender |
|---|---|---|---|---|---|---|---|
| control | 58559 | 39.06 | 38.8 | 39.33 | 0 | 999 | 0.4885 |
| test | 42682 | 13.75 | 13.64 | 13.85 | 0 | 52 | 0.5104 |
## Ttest
tidy(t.test(birthorder$age ~ birthorder$group_birthorder, var.equal = T))| estimate1 | estimate2 | statistic | p.value | parameter | conf.low | conf.high | method | alternative |
|---|---|---|---|---|---|---|---|---|
| 39.06 | 13.75 | 114.2 | 0 | 69280 | 24.89 | 25.75 | Two Sample t-test | two.sided |
cohen.d(birthorder$age, as.factor(birthorder$group_birthorder), na.rm = T)##
## Cohen's d
##
## d estimate: 0.9244 (large)
## 95 percent confidence interval:
## lower upper
## 0.9078 0.9410gender = birthorder %>%
group_by(group_birthorder) %>%
summarise(gender = sum(male == 1, na.rm=T),
gender2 = sum(male ==0,na.rm=T)) %>%
select(gender, gender2)
prop.table(gender)| gender | gender2 |
|---|---|
| 0.3284 | 0.3438 |
| 0.1673 | 0.1605 |
tidy(chisq.test(gender))| statistic | p.value | parameter | method |
|---|---|---|---|
| 29.15 | 0.0000000671 | 1 | Pearson’s Chi-squared test with Yates’ continuity correction |
## Ratings
ratings = birthorder %>%
group_by(group_birthorder) %>%
summarise(g_factor_mean = mean(g_factor_2015_old, na.rm=T),
g_factor_confidence_low= mean(g_factor_2015_old, na.rm = T) -
(qt(.975, n()-1)*sd(g_factor_2015_old, na.rm = T)/sqrt(n())),
g_factor_confidence_high = mean(g_factor_2015_old, na.rm = T) +
(qt(.975, n()-1)*sd(g_factor_2015_old, na.rm = T)/sqrt(n())),
big5_ext_mean = mean(big5_ext, na.rm=T),
big5_ext_confidence_low= mean(big5_ext, na.rm = T) -
(qt(.975, n()-1)*sd(big5_ext, na.rm = T)/sqrt(n())),
big5_ext_confidence_high = mean(big5_ext, na.rm = T) +
(qt(.975, n()-1)*sd(big5_ext, na.rm = T)/sqrt(n())),
big5_neu_mean = mean(big5_neu, na.rm=T),
big5_neu_confidence_low= mean(big5_neu, na.rm = T) -
(qt(.975, n()-1)*sd(big5_neu, na.rm = T)/sqrt(n())),
big5_neu_confidence_high = mean(big5_neu, na.rm = T) +
(qt(.975, n()-1)*sd(big5_neu, na.rm = T)/sqrt(n())),
big5_con_mean = mean(big5_con, na.rm=T),
big5_con_confidence_low= mean(big5_con, na.rm = T) -
(qt(.975, n()-1)*sd(big5_con, na.rm = T)/sqrt(n())),
big5_con_confidence_high = mean(big5_con, na.rm = T) +
(qt(.975, n()-1)*sd(big5_con, na.rm = T)/sqrt(n())),
big5_agree_mean = mean(big5_agree, na.rm=T),
big5_agree_confidence_low= mean(big5_agree, na.rm = T) -
(qt(.975, n()-1)*sd(big5_agree, na.rm = T)/sqrt(n())),
big5_agree_confidence_high = mean(big5_agree, na.rm = T) +
(qt(.975, n()-1)*sd(big5_agree, na.rm = T)/sqrt(n())),
big5_open_mean = mean(big5_open, na.rm=T),
big5_open_confidence_low= mean(big5_open, na.rm = T) -
(qt(.975, n()-1)*sd(big5_open, na.rm = T)/sqrt(n())),
big5_open_confidence_high = mean(big5_open, na.rm = T) +
(qt(.975, n()-1)*sd(big5_open, na.rm = T)/sqrt(n())),
riskA_mean = mean(riskA, na.rm=T),
riskA_confidence_low= mean(riskA, na.rm = T) -
(qt(.975, n()-1)*sd(riskA, na.rm = T)/sqrt(n())),
riskA_confidence_high = mean(riskA, na.rm = T) +
(qt(.975, n()-1)*sd(riskA, na.rm = T)/sqrt(n())),
riskB_mean = mean(riskB, na.rm=T),
riskB_confidence_low= mean(riskB, na.rm = T) -
(qt(.975, n()-1)*sd(riskB, na.rm = T)/sqrt(n())),
riskB_confidence_high = mean(riskB, na.rm = T) +
(qt(.975, n()-1)*sd(riskB, na.rm = T)/sqrt(n())))
ratings| group_birthorder | g_factor_mean | g_factor_confidence_low | g_factor_confidence_high | big5_ext_mean | big5_ext_confidence_low | big5_ext_confidence_high | big5_neu_mean | big5_neu_confidence_low | big5_neu_confidence_high | big5_con_mean | big5_con_confidence_low | big5_con_confidence_high | big5_agree_mean | big5_agree_confidence_low | big5_agree_confidence_high | big5_open_mean | big5_open_confidence_low | big5_open_confidence_high | riskA_mean | riskA_confidence_low | riskA_confidence_high | riskB_mean | riskB_confidence_low | riskB_confidence_high |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| control | -0.1381 | -0.1448 | -0.1314 | 3.429 | 3.424 | 3.434 | 2.669 | 2.664 | 2.674 | 3.837 | 3.833 | 3.842 | 3.915 | 3.911 | 3.919 | 3.674 | 3.668 | 3.68 | 3.433 | 3.421 | 3.446 | 4.185 | 4.174 | 4.197 |
| test | 0.4502 | 0.4438 | 0.4567 | 3.493 | 3.487 | 3.5 | 2.724 | 2.717 | 2.73 | 3.73 | 3.724 | 3.735 | 3.85 | 3.846 | 3.855 | 3.817 | 3.812 | 3.823 | 3.292 | 3.278 | 3.305 | 4.288 | 4.276 | 4.301 |
tidy(t.test(birthorder$g_factor_2015_old ~ birthorder$group_birthorder, var.equal = T))| estimate1 | estimate2 | statistic | p.value | parameter | conf.low | conf.high | method | alternative |
|---|---|---|---|---|---|---|---|---|
| -0.1381 | 0.4502 | -51.98 | 0 | 27524 | -0.6105 | -0.5661 | Two Sample t-test | two.sided |
cohen.d(birthorder$g_factor_2015_old, as.factor(birthorder$group_birthorder), na.rm = T)##
## Cohen's d
##
## d estimate: -0.7392 (medium)
## 95 percent confidence interval:
## lower upper
## -0.7678 -0.7107tidy(t.test(birthorder$years_of_education ~ birthorder$group_birthorder, var.equal = T))| estimate1 | estimate2 | statistic | p.value | parameter | conf.low | conf.high | method | alternative |
|---|---|---|---|---|---|---|---|---|
| 8.283 | 11.4 | -49.98 | 0 | 33816 | -3.236 | -2.992 | Two Sample t-test | two.sided |
cohen.d(birthorder$years_of_education, as.factor(birthorder$group_birthorder), na.rm = T)##
## Cohen's d
##
## d estimate: -0.6763 (medium)
## 95 percent confidence interval:
## lower upper
## -0.7033 -0.6493tidy(t.test(birthorder$big5_ext ~ birthorder$group_birthorder, var.equal = T))| estimate1 | estimate2 | statistic | p.value | parameter | conf.low | conf.high | method | alternative |
|---|---|---|---|---|---|---|---|---|
| 3.429 | 3.493 | -6.982 | 2.971e-12 | 31444 | -0.08258 | -0.04638 | Two Sample t-test | two.sided |
cohen.d(birthorder$big5_ext, as.factor(birthorder$group_birthorder), na.rm = T)##
## Cohen's d
##
## d estimate: -0.09677 (negligible)
## 95 percent confidence interval:
## lower upper
## -0.12395 -0.06959tidy(t.test(birthorder$big5_neu ~ birthorder$group_birthorder, var.equal = T))| estimate1 | estimate2 | statistic | p.value | parameter | conf.low | conf.high | method | alternative |
|---|---|---|---|---|---|---|---|---|
| 2.669 | 2.724 | -5.923 | 0.000000003185 | 31444 | -0.07268 | -0.03654 | Two Sample t-test | two.sided |
cohen.d(birthorder$big5_neu, as.factor(birthorder$group_birthorder), na.rm = T)##
## Cohen's d
##
## d estimate: -0.0821 (negligible)
## 95 percent confidence interval:
## lower upper
## -0.10927 -0.05493tidy(t.test(birthorder$big5_con ~ birthorder$group_birthorder, var.equal = T))| estimate1 | estimate2 | statistic | p.value | parameter | conf.low | conf.high | method | alternative |
|---|---|---|---|---|---|---|---|---|
| 3.837 | 3.73 | 14.06 | 9.458e-45 | 31444 | 0.09249 | 0.1225 | Two Sample t-test | two.sided |
cohen.d(birthorder$big5_con, as.factor(birthorder$group_birthorder), na.rm = T)##
## Cohen's d
##
## d estimate: 0.1948 (negligible)
## 95 percent confidence interval:
## lower upper
## 0.1676 0.2221tidy(t.test(birthorder$big5_agree ~ birthorder$group_birthorder, var.equal = T))| estimate1 | estimate2 | statistic | p.value | parameter | conf.low | conf.high | method | alternative |
|---|---|---|---|---|---|---|---|---|
| 3.915 | 3.85 | 9.136 | 6.863e-20 | 31444 | 0.0508 | 0.07854 | Two Sample t-test | two.sided |
cohen.d(birthorder$big5_agree, as.factor(birthorder$group_birthorder), na.rm = T)##
## Cohen's d
##
## d estimate: 0.1266 (negligible)
## 95 percent confidence interval:
## lower upper
## 0.09944 0.15381tidy(t.test(birthorder$big5_open ~ birthorder$group_birthorder, var.equal = T))| estimate1 | estimate2 | statistic | p.value | parameter | conf.low | conf.high | method | alternative |
|---|---|---|---|---|---|---|---|---|
| 3.674 | 3.817 | -15.52 | 3.76e-54 | 31444 | -0.1615 | -0.1253 | Two Sample t-test | two.sided |
cohen.d(birthorder$big5_open, as.factor(birthorder$group_birthorder), na.rm = T)##
## Cohen's d
##
## d estimate: -0.2152 (small)
## 95 percent confidence interval:
## lower upper
## -0.2424 -0.1880tidy(t.test(birthorder$riskA ~ birthorder$group_birthorder, var.equal = T))| estimate1 | estimate2 | statistic | p.value | parameter | conf.low | conf.high | method | alternative |
|---|---|---|---|---|---|---|---|---|
| 3.433 | 3.292 | 6.415 | 0.0000000001434 | 27778 | 0.0985 | 0.1852 | Two Sample t-test | two.sided |
cohen.d(birthorder$riskA, as.factor(birthorder$group_birthorder), na.rm = T)##
## Cohen's d
##
## d estimate: 0.094 (negligible)
## 95 percent confidence interval:
## lower upper
## 0.06526 0.12273tidy(t.test(birthorder$riskB ~ birthorder$group_birthorder, var.equal = T))| estimate1 | estimate2 | statistic | p.value | parameter | conf.low | conf.high | method | alternative |
|---|---|---|---|---|---|---|---|---|
| 4.185 | 4.288 | -5.116 | 0.0000003147 | 29576 | -0.1426 | -0.06362 | Two Sample t-test | two.sided |
cohen.d(birthorder$riskB, as.factor(birthorder$group_birthorder), na.rm = T)##
## Cohen's d
##
## d estimate: -0.0729 (negligible)
## 95 percent confidence interval:
## lower upper
## -0.10084 -0.04496## Educational Attainment
educational_attainment = birthorder %>%
group_by(group_birthorder) %>%
summarise(years_of_education_mean = mean(years_of_education, na.rm=T),
years_of_education_confidence_low= mean(years_of_education, na.rm = T) -
(qt(.975, n()-1)*sd(years_of_education, na.rm = T)/sqrt(n())),
years_of_education_high = mean(years_of_education, na.rm = T) +
(qt(.975, n()-1)*sd(years_of_education, na.rm = T)/sqrt(n())))
educational_attainment| group_birthorder | years_of_education_mean | years_of_education_confidence_low | years_of_education_high |
|---|---|---|---|
| control | 8.283 | 8.244 | 8.322 |
| test | 11.4 | 11.36 | 11.43 |
t.test(birthorder$years_of_education ~ birthorder$group_birthorder, var.equal = T)| Test statistic | df | P value | Alternative hypothesis | mean in group control | mean in group test |
|---|---|---|---|---|---|
| -49.98 | 33816 | 0 * * * | two.sided | 8.283 | 11.4 |
ggplot(data=birthorder, aes(x=years_of_education, fill=group_birthorder)) +
geom_histogram(stat="count", binwidth=.5, position="dodge")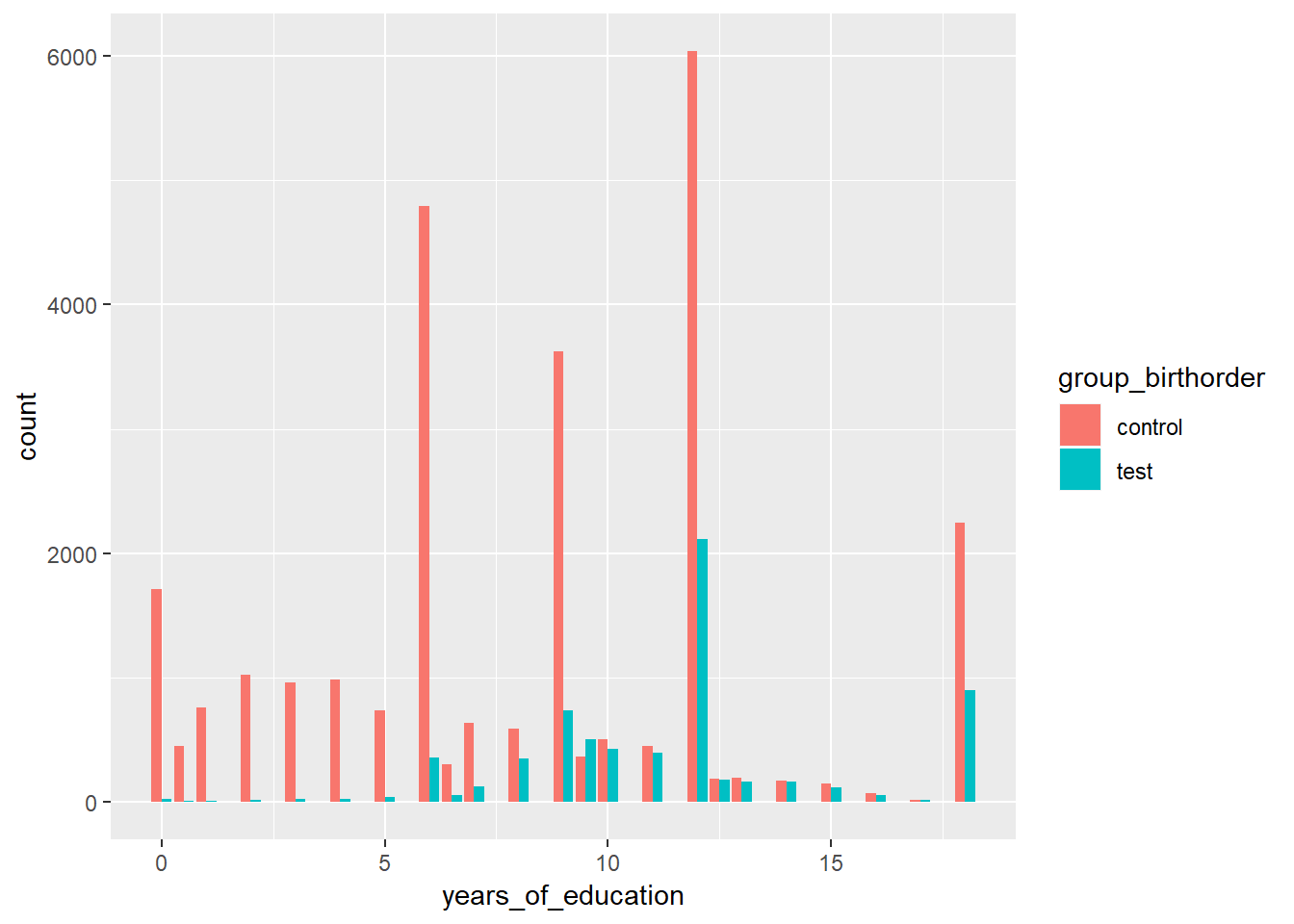
Descriptives
x = birthorder %>% filter(group_birthorder == "test", group_outcome == "test")
mean_sd = birthorder %>%
group_by(group_birthorder) %>%
summarise(age_mean = mean(age, na.rm=T),
age_sd = sd(age, na.rm=T),
g_factor_mean = mean(g_factor_2015_old, na.rm=T),
g_factor_sd = sd(g_factor_2015_old, na.rm=T),
big5_ext_mean = mean(big5_ext, na.rm=T),
big5_ext_sd = sd(big5_ext, na.rm=T),
big5_neu_mean = mean(big5_neu, na.rm=T),
big5_neu_sd = sd(big5_neu, na.rm=T),
big5_con_mean = mean(big5_con, na.rm=T),
big5_con_sd = sd(big5_con, na.rm=T),
big5_agree_mean = mean(big5_agree, na.rm=T),
big5_agree_sd = sd(big5_agree, na.rm=T),
big5_open_mean = mean(big5_open, na.rm=T),
big5_open_sd = sd(big5_open, na.rm=T),
riskA_mean = mean(riskA, na.rm=T),
riskA_sd = sd(riskA, na.rm=T),
riskB_mean = mean(riskB, na.rm=T),
riskB_sd = sd(riskB, na.rm=T),
years_of_education_mean = mean(years_of_education, na.rm=T),
years_of_education_sd = sd(years_of_education, na.rm=T))
mean_sd| group_birthorder | age_mean | age_sd | g_factor_mean | g_factor_sd | big5_ext_mean | big5_ext_sd | big5_neu_mean | big5_neu_sd | big5_con_mean | big5_con_sd | big5_agree_mean | big5_agree_sd | big5_open_mean | big5_open_sd | riskA_mean | riskA_sd | riskB_mean | riskB_sd | years_of_education_mean | years_of_education_sd |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| control | 39.06 | 32.54 | -0.1381 | 0.8272 | 3.429 | 0.6616 | 2.669 | 0.6655 | 3.837 | 0.5447 | 3.915 | 0.5111 | 3.674 | 0.6824 | 3.433 | 1.528 | 4.185 | 1.441 | 8.283 | 4.847 |
| test | 13.75 | 10.83 | 0.4502 | 0.6835 | 3.493 | 0.6839 | 2.724 | 0.6638 | 3.73 | 0.5768 | 3.85 | 0.5094 | 3.817 | 0.6013 | 3.292 | 1.437 | 4.288 | 1.313 | 11.4 | 3.488 |
Correlations
cor = round(cor(birthorder %>% filter(group_birthorder == "test", group_outcome == "test") %>% ungroup() %>% select(age, male, g_factor_2015_old, years_of_education, big5_ext, big5_neu, big5_con, big5_agree, big5_open, riskA, riskB), use = "pairwise.complete.obs"), 2)
cor| age | male | g_factor_2015_old | years_of_education | big5_ext | big5_neu | big5_con | big5_agree | big5_open | riskA | riskB |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | -0.03 | -0.1 | 0.24 | 0 | -0.13 | 0.24 | 0.1 | 0 | -0.06 | -0.01 |
| -0.03 | 1 | -0.01 | -0.05 | -0.13 | -0.13 | 0.02 | 0.03 | 0.08 | -0.13 | -0.12 |
| -0.1 | -0.01 | 1 | 0.35 | 0.06 | -0.05 | -0.03 | -0.04 | 0.08 | -0.15 | 0.05 |
| 0.24 | -0.05 | 0.35 | 1 | 0.07 | -0.08 | 0.1 | 0.03 | 0.15 | -0.19 | -0.02 |
| 0 | -0.13 | 0.06 | 0.07 | 1 | -0.09 | 0.07 | 0.07 | 0.17 | -0.01 | 0 |
| -0.13 | -0.13 | -0.05 | -0.08 | -0.09 | 1 | -0.2 | -0.17 | -0.07 | 0.04 | 0.02 |
| 0.24 | 0.02 | -0.03 | 0.1 | 0.07 | -0.2 | 1 | 0.32 | 0.27 | -0.04 | 0.02 |
| 0.1 | 0.03 | -0.04 | 0.03 | 0.07 | -0.17 | 0.32 | 1 | 0.23 | -0.02 | -0.01 |
| 0 | 0.08 | 0.08 | 0.15 | 0.17 | -0.07 | 0.27 | 0.23 | 1 | -0.08 | -0.03 |
| -0.06 | -0.13 | -0.15 | -0.19 | -0.01 | 0.04 | -0.04 | -0.02 | -0.08 | 1 | 0.3 |
| -0.01 | -0.12 | 0.05 | -0.02 | 0 | 0.02 | 0.02 | -0.01 | -0.03 | 0.3 | 1 |
Reliability
Intelligence
alpha(birthorder %>% filter(group_birthorder == "test", group_outcome == "test") %>% ungroup() %>% select(raven_2015_old, math_2015_old, count_backwards, words_delayed, adaptive_numbering))##
## Reliability analysis
## Call: alpha(x = birthorder %>% filter(group_birthorder == "test", group_outcome ==
## "test") %>% ungroup() %>% select(raven_2015_old, math_2015_old,
## count_backwards, words_delayed, adaptive_numbering))
##
## raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
## 0.025 0.61 0.56 0.24 1.6 0.00066 109 16 0.22
##
## lower alpha upper 95% confidence boundaries
## 0.02 0.03 0.03
##
## Reliability if an item is dropped:
## raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r
## raven_2015_old 0.024 0.54 0.47 0.23 1.2 0.00069 0.0026 0.22
## math_2015_old 0.022 0.53 0.46 0.22 1.1 0.00068 0.0029 0.19
## count_backwards 0.023 0.57 0.51 0.25 1.4 0.00069 0.0042 0.25
## words_delayed 0.011 0.60 0.53 0.27 1.5 0.00020 0.0027 0.29
## adaptive_numbering 0.214 0.53 0.46 0.22 1.1 0.00484 0.0040 0.20
##
## Item statistics
## n raw.r std.r r.cor r.drop mean sd
## raven_2015_old 6598 0.27 0.64 0.50 0.30 0.75 0.21
## math_2015_old 6598 0.26 0.66 0.54 0.30 0.44 0.30
## count_backwards 6492 0.26 0.60 0.42 0.28 0.80 0.26
## words_delayed 6593 0.20 0.56 0.35 0.20 5.04 1.68
## adaptive_numbering 6581 0.88 0.66 0.53 0.29 539.96 54.83Personality
##Extraversion
alpha(birthorder %>% filter(group_birthorder == "test", group_outcome == "test") %>% ungroup() %>%
select(e1, e2r_reversed, e3))##
## Reliability analysis
## Call: alpha(x = birthorder %>% filter(group_birthorder == "test", group_outcome ==
## "test") %>% ungroup() %>% select(e1, e2r_reversed, e3))
##
## raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
## 0.45 0.43 0.35 0.2 0.77 0.0072 3.5 0.68 0.17
##
## lower alpha upper 95% confidence boundaries
## 0.43 0.45 0.46
##
## Reliability if an item is dropped:
## raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r
## e1 0.17 0.19 0.11 0.11 0.24 0.0121 NA 0.11
## e2r_reversed 0.26 0.29 0.17 0.17 0.42 0.0106 NA 0.17
## e3 0.50 0.50 0.33 0.33 1.00 0.0083 NA 0.33
##
## Item statistics
## n raw.r std.r r.cor r.drop mean sd
## e1 6584 0.79 0.73 0.52 0.36 3.2 1.13
## e2r_reversed 6584 0.76 0.70 0.45 0.32 3.0 1.11
## e3 6584 0.48 0.62 0.25 0.17 4.2 0.67
##
## Non missing response frequency for each item
## 1 2 3 4 5 miss
## e1 0.02 0.35 0.11 0.40 0.12 0.55
## e2r_reversed 0.07 0.34 0.11 0.43 0.05 0.55
## e3 0.00 0.02 0.06 0.61 0.30 0.55## Neuroticism
alpha(birthorder %>% filter(group_birthorder == "test", group_outcome == "test") %>% ungroup()
%>% select(n1r_reversed, n2, n3))##
## Reliability analysis
## Call: alpha(x = birthorder %>% filter(group_birthorder == "test", group_outcome ==
## "test") %>% ungroup() %>% select(n1r_reversed, n2, n3))
##
## raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
## 0.37 0.33 0.29 0.14 0.5 0.0084 2.7 0.66 0.056
##
## lower alpha upper 95% confidence boundaries
## 0.35 0.37 0.38
##
## Reliability if an item is dropped:
## raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r
## n1r_reversed 0.510 0.510 0.342 0.342 1.041 0.0081 NA 0.342
## n2 0.099 0.105 0.056 0.056 0.118 0.0140 NA 0.056
## n3 0.059 0.062 0.032 0.032 0.067 0.0145 NA 0.032
##
## Item statistics
## n raw.r std.r r.cor r.drop mean sd
## n1r_reversed 6584 0.43 0.55 0.087 0.053 2.1 0.77
## n2 6584 0.75 0.70 0.469 0.291 3.1 1.11
## n3 6584 0.76 0.71 0.493 0.308 3.0 1.09
##
## Non missing response frequency for each item
## 1 2 3 4 5 miss
## n1r_reversed 0.17 0.67 0.08 0.07 0.01 0.55
## n2 0.02 0.39 0.10 0.40 0.09 0.55
## n3 0.03 0.46 0.09 0.36 0.06 0.55##conscientiousness
alpha(birthorder %>% filter(group_birthorder == "test", group_outcome == "test") %>% ungroup() %>% select(c1, c2r_reversed, c3))##
## Reliability analysis
## Call: alpha(x = birthorder %>% filter(group_birthorder == "test", group_outcome ==
## "test") %>% ungroup() %>% select(c1, c2r_reversed, c3))
##
## raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
## 0.37 0.39 0.31 0.18 0.65 0.009 3.7 0.58 0.19
##
## lower alpha upper 95% confidence boundaries
## 0.36 0.37 0.39
##
## Reliability if an item is dropped:
## raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r
## c1 0.17 0.17 0.094 0.094 0.21 0.0135 NA 0.094
## c2r_reversed 0.39 0.40 0.249 0.249 0.66 0.0098 NA 0.249
## c3 0.31 0.32 0.189 0.189 0.47 0.0109 NA 0.189
##
## Item statistics
## n raw.r std.r r.cor r.drop mean sd
## c1 6584 0.65 0.71 0.47 0.29 4.1 0.73
## c2r_reversed 6584 0.70 0.64 0.29 0.17 3.4 0.99
## c3 6584 0.65 0.67 0.36 0.21 3.8 0.86
##
## Non missing response frequency for each item
## 1 2 3 4 5 miss
## c1 0.00 0.04 0.07 0.63 0.25 0.55
## c2r_reversed 0.03 0.25 0.11 0.56 0.05 0.55
## c3 0.01 0.11 0.12 0.63 0.13 0.55##Agreeableness
alpha(birthorder %>% filter(group_birthorder == "test", group_outcome == "test") %>% ungroup() %>% select(a1, a2, a3r_reversed))##
## Reliability analysis
## Call: alpha(x = birthorder %>% filter(group_birthorder == "test", group_outcome ==
## "test") %>% ungroup() %>% select(a1, a2, a3r_reversed))
##
## raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
## 0.28 0.36 0.33 0.16 0.57 0.01 3.9 0.51 0.047
##
## lower alpha upper 95% confidence boundaries
## 0.26 0.28 0.3
##
## Reliability if an item is dropped:
## raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r
## a1 0.048 0.054 0.028 0.028 0.057 0.014 NA 0.028
## a2 0.081 0.089 0.047 0.047 0.098 0.014 NA 0.047
## a3r_reversed 0.577 0.577 0.406 0.406 1.367 0.007 NA 0.406
##
## Item statistics
## n raw.r std.r r.cor r.drop mean sd
## a1 6584 0.63 0.73 0.544 0.250 4.2 0.66
## a2 6584 0.61 0.72 0.526 0.236 4.1 0.64
## a3r_reversed 6584 0.71 0.54 0.067 0.044 3.2 1.03
##
## Non missing response frequency for each item
## 1 2 3 4 5 miss
## a1 0.00 0.02 0.06 0.62 0.30 0.55
## a2 0.00 0.02 0.08 0.66 0.24 0.55
## a3r_reversed 0.03 0.29 0.12 0.51 0.05 0.55##Openness
alpha(birthorder %>% filter(group_birthorder == "test", group_outcome == "test") %>% ungroup() %>% select(o1, o2, o3))##
## Reliability analysis
## Call: alpha(x = birthorder %>% filter(group_birthorder == "test", group_outcome ==
## "test") %>% ungroup() %>% select(o1, o2, o3))
##
## raw_alpha std.alpha G6(smc) average_r S/N ase mean sd median_r
## 0.46 0.46 0.37 0.22 0.86 0.0077 3.8 0.6 0.22
##
## lower alpha upper 95% confidence boundaries
## 0.44 0.46 0.47
##
## Reliability if an item is dropped:
## raw_alpha std.alpha G6(smc) average_r S/N alpha se var.r med.r
## o1 0.32 0.33 0.20 0.20 0.49 0.0109 NA 0.20
## o2 0.40 0.40 0.25 0.25 0.67 0.0098 NA 0.25
## o3 0.36 0.36 0.22 0.22 0.57 0.0104 NA 0.22
##
## Item statistics
## n raw.r std.r r.cor r.drop mean sd
## o1 6584 0.71 0.71 0.45 0.30 3.8 0.87
## o2 6584 0.72 0.68 0.39 0.27 3.6 0.96
## o3 6584 0.65 0.69 0.42 0.28 4.1 0.76
##
## Non missing response frequency for each item
## 1 2 3 4 5 miss
## o1 0.01 0.12 0.11 0.62 0.14 0.55
## o2 0.02 0.17 0.13 0.56 0.13 0.55
## o3 0.01 0.05 0.07 0.61 0.27 0.55Plots age and gender
Plots Gender
birthorder = birthorder %>% filter(age<=100)
birthorder = birthorder %>%
mutate(outcome = age)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = birthorder_genes)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = sibling_count_genes)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = g_factor_2015_old)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = g_factor_2015_young)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = g_factor_2007_old)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = g_factor_2007_young)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = big5_ext)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = big5_con)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = big5_open)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = big5_neu)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = big5_agree)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = riskA)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = riskB)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = years_of_education)
plot_gender(birthorder)
plot_gender(birthorder %>% filter(!is.na(attended_school)) %>% mutate(outcome = as.numeric(attended_school)))
birthorder = birthorder %>%
mutate(outcome = Elementary_missed)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = Elementary_worked)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = wage_last_month_log)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = wage_last_year_log)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = Self_employed)
plot_gender(birthorder)
birthorder = birthorder %>%
mutate(outcome = ever_smoked)
plot_gender(birthorder)
Plots Age
plot_age(birthorder %>% filter(age<= 100) %>% mutate(outcome = birthorder_genes))## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'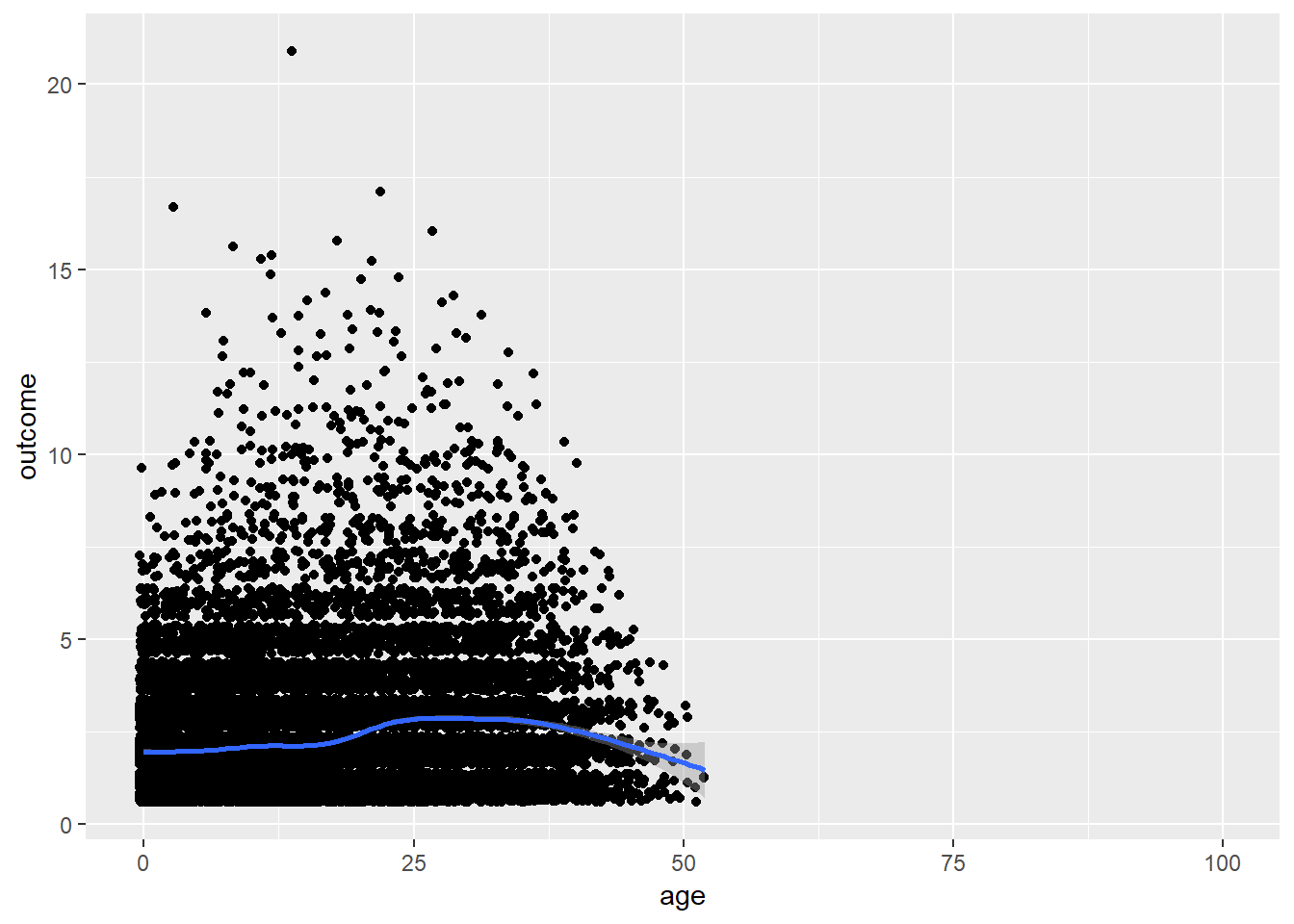
birthorder = birthorder %>%
mutate(outcome = sibling_count_genes)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'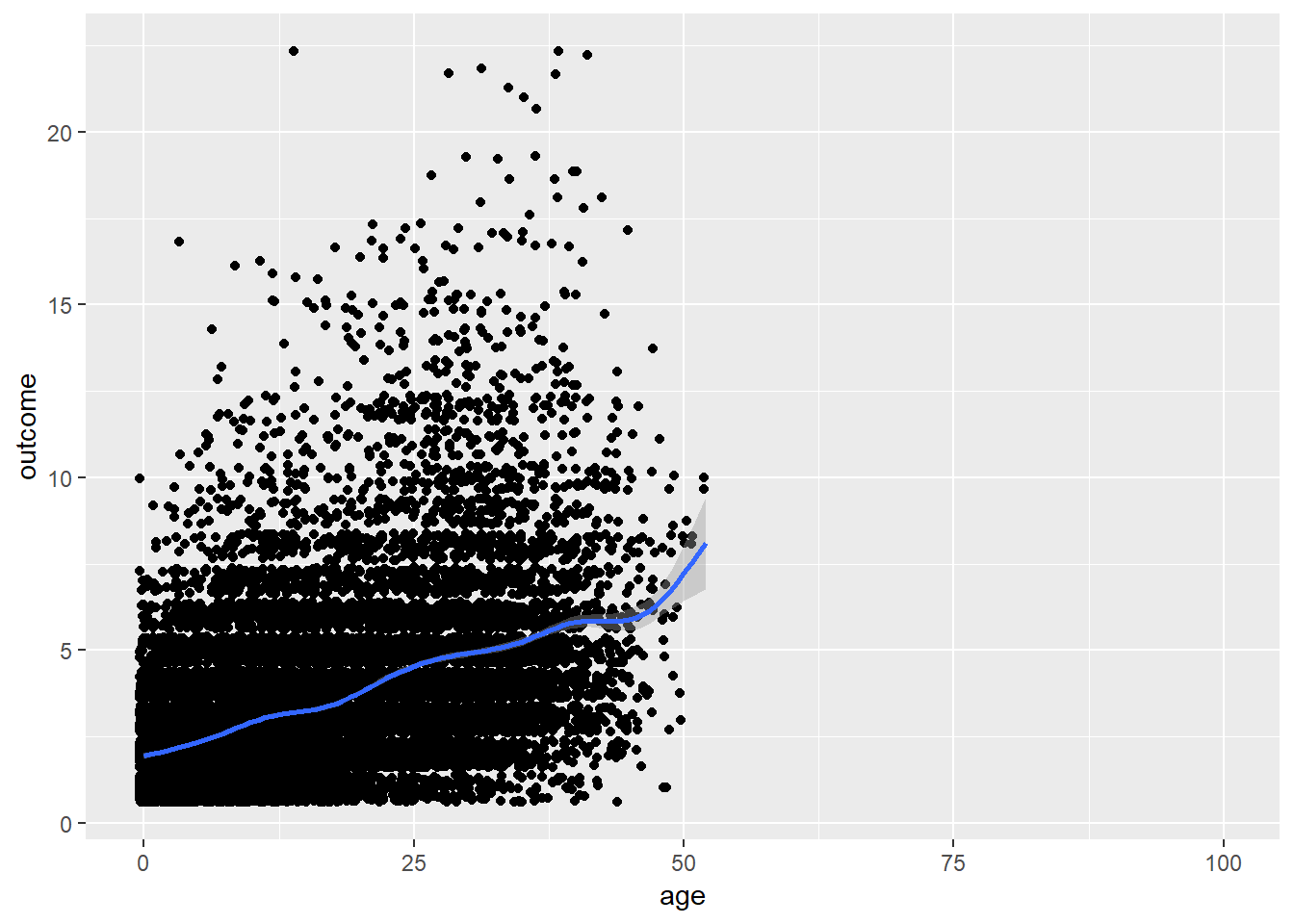
birthorder = birthorder %>%
mutate(outcome = g_factor_2015_old)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'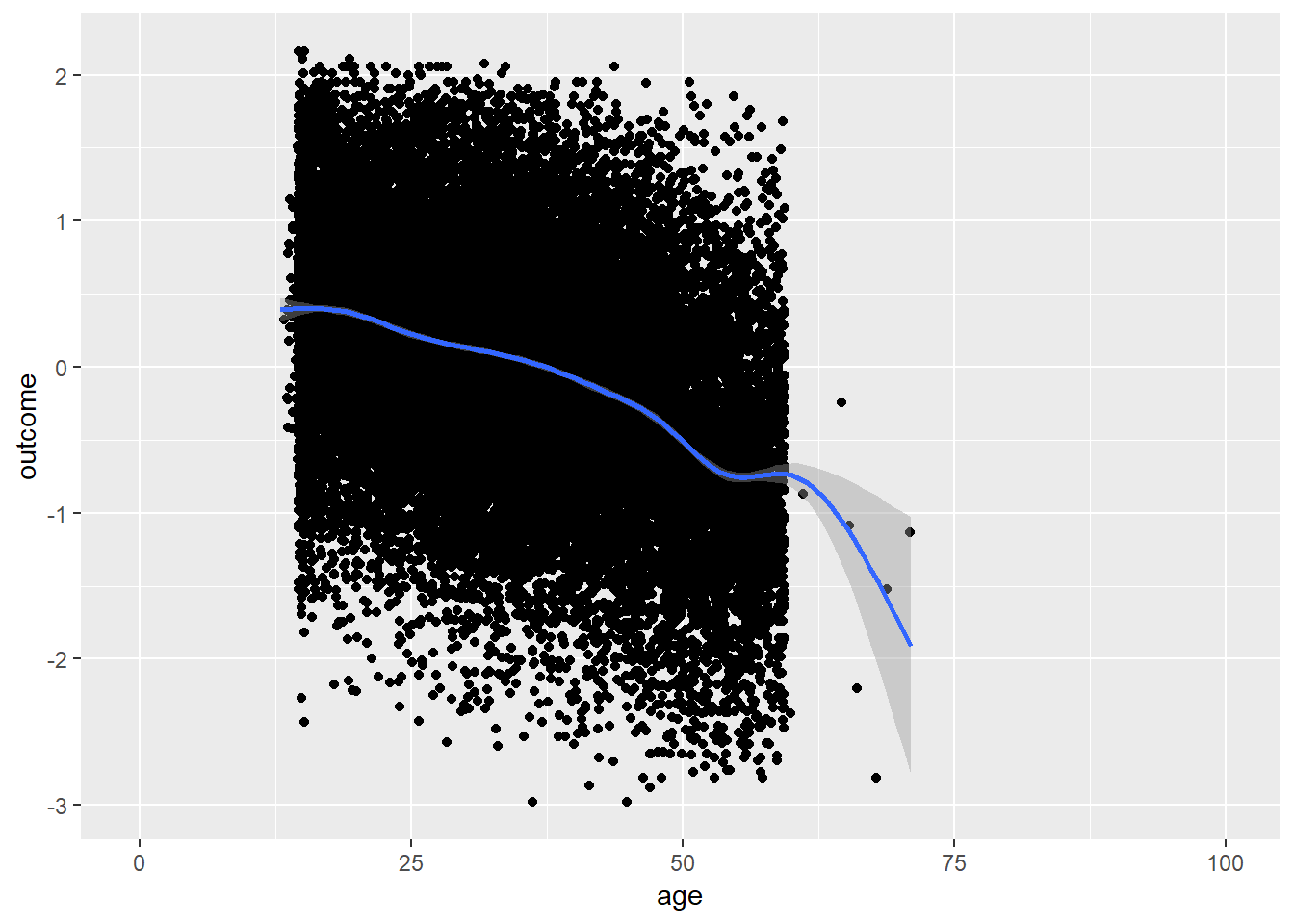
birthorder = birthorder %>%
mutate(outcome = g_factor_2015_young)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'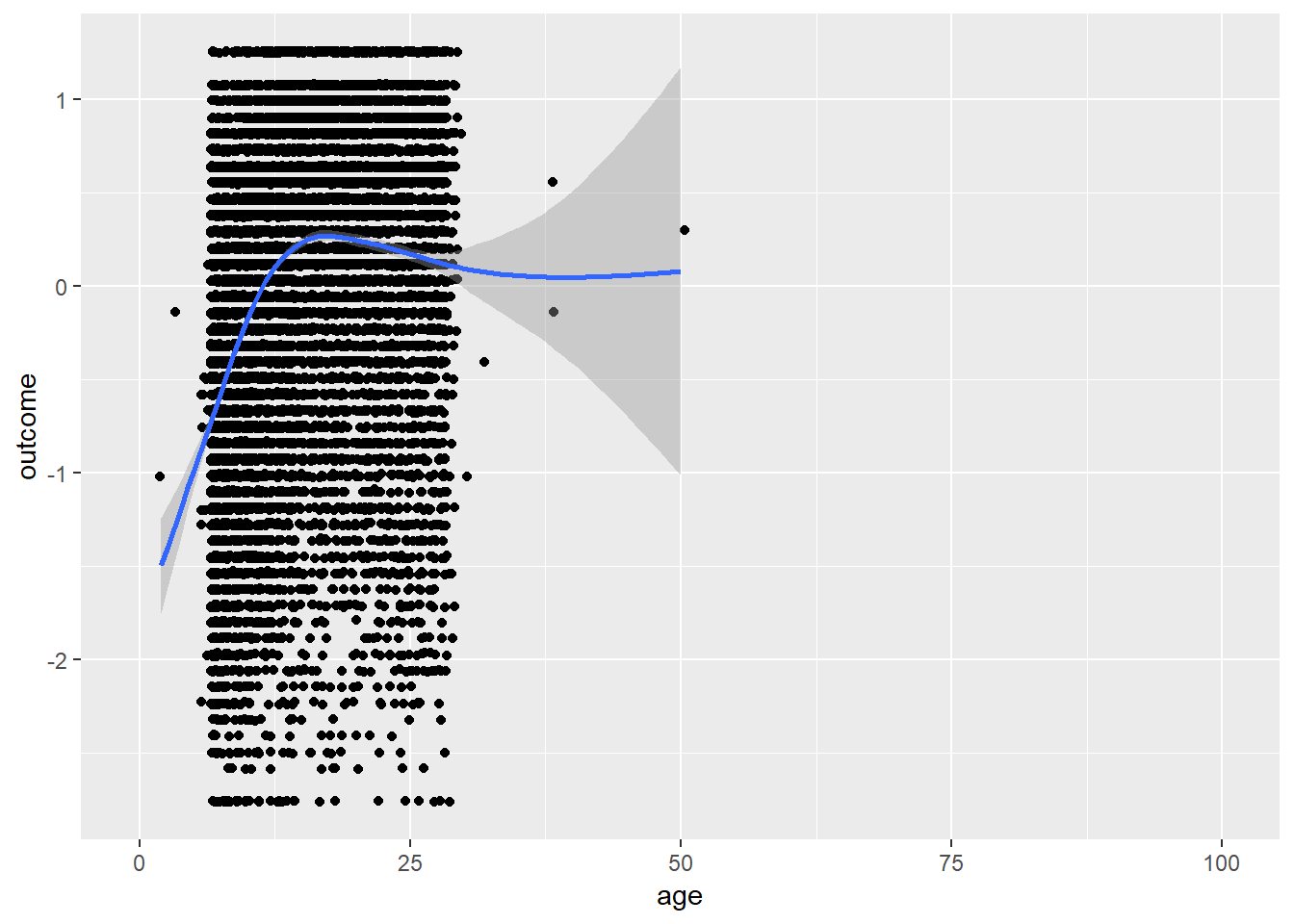
birthorder = birthorder %>%
mutate(outcome = g_factor_2007_old)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'
birthorder = birthorder %>%
mutate(outcome = g_factor_2007_young)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'
birthorder = birthorder %>%
mutate(outcome = big5_ext)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'
birthorder = birthorder %>%
mutate(outcome = big5_con)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'
birthorder = birthorder %>%
mutate(outcome = big5_open)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'
birthorder = birthorder %>%
mutate(outcome = big5_neu)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'
birthorder = birthorder %>%
mutate(outcome = big5_agree)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'
birthorder = birthorder %>%
mutate(outcome = riskA)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'
birthorder = birthorder %>%
mutate(outcome = riskB)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'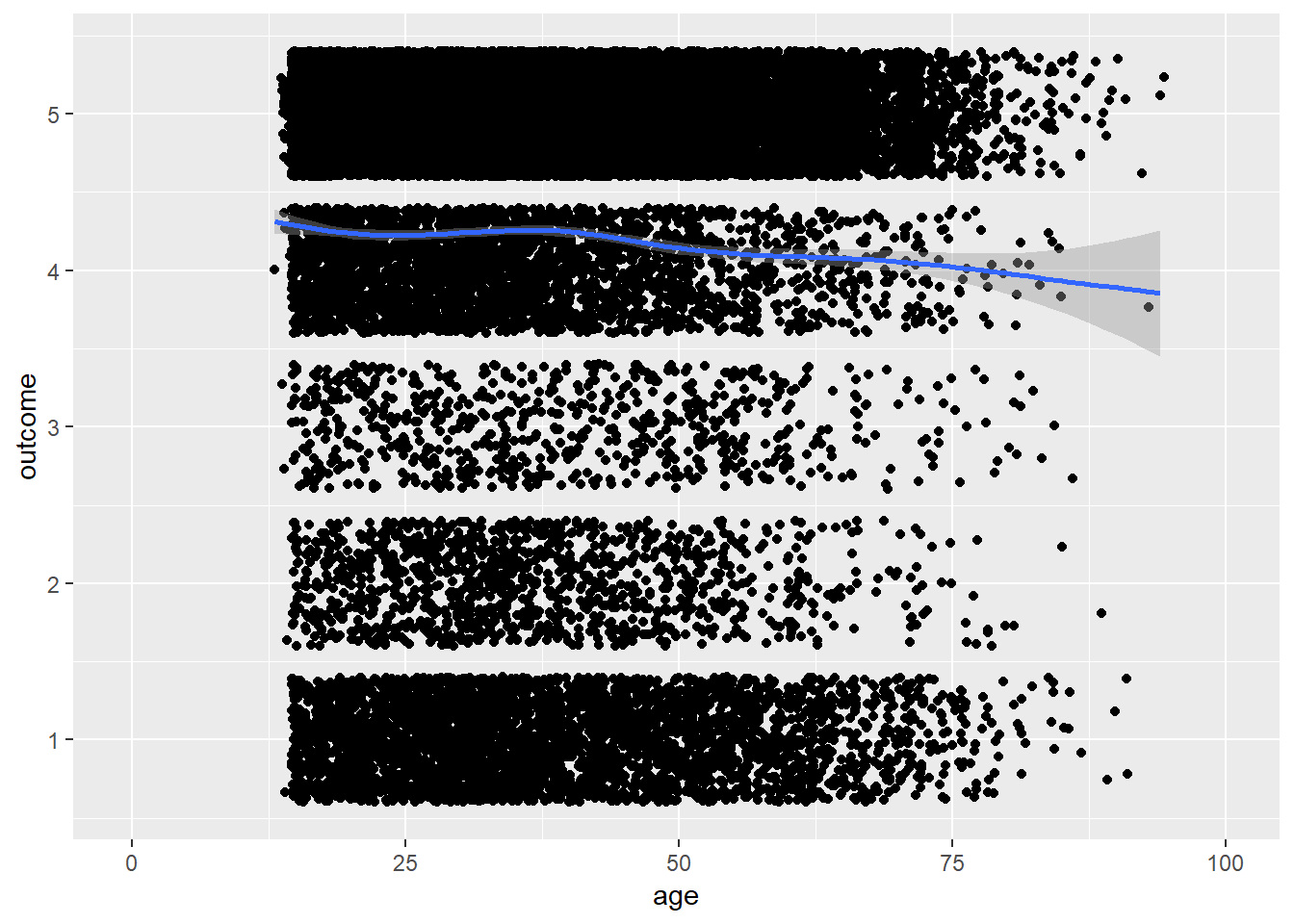
birthorder = birthorder %>%
mutate(outcome = years_of_education)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'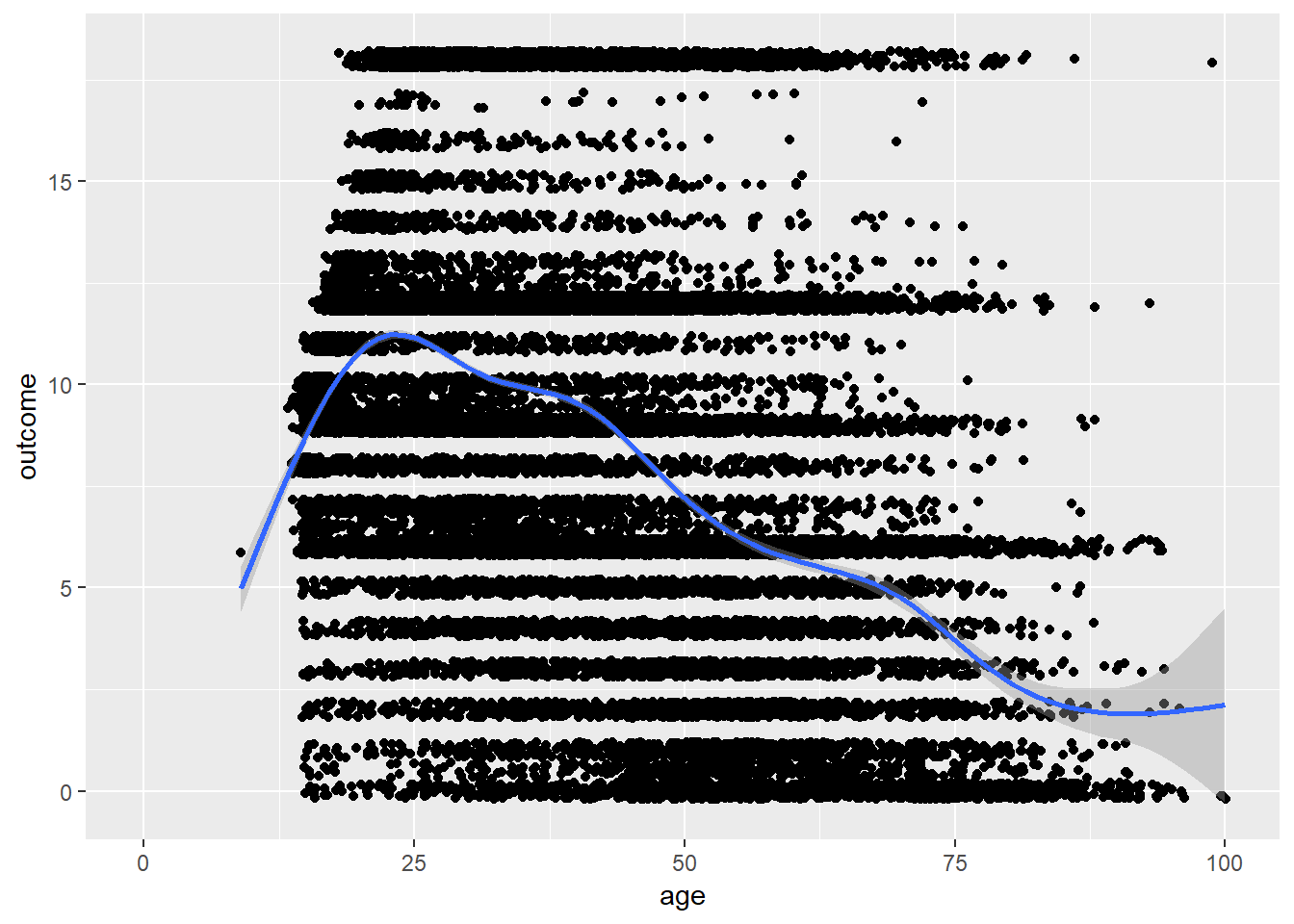
plot_age(birthorder %>% filter(!is.na(attended_school)) %>% mutate(outcome = as.numeric(attended_school)))## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'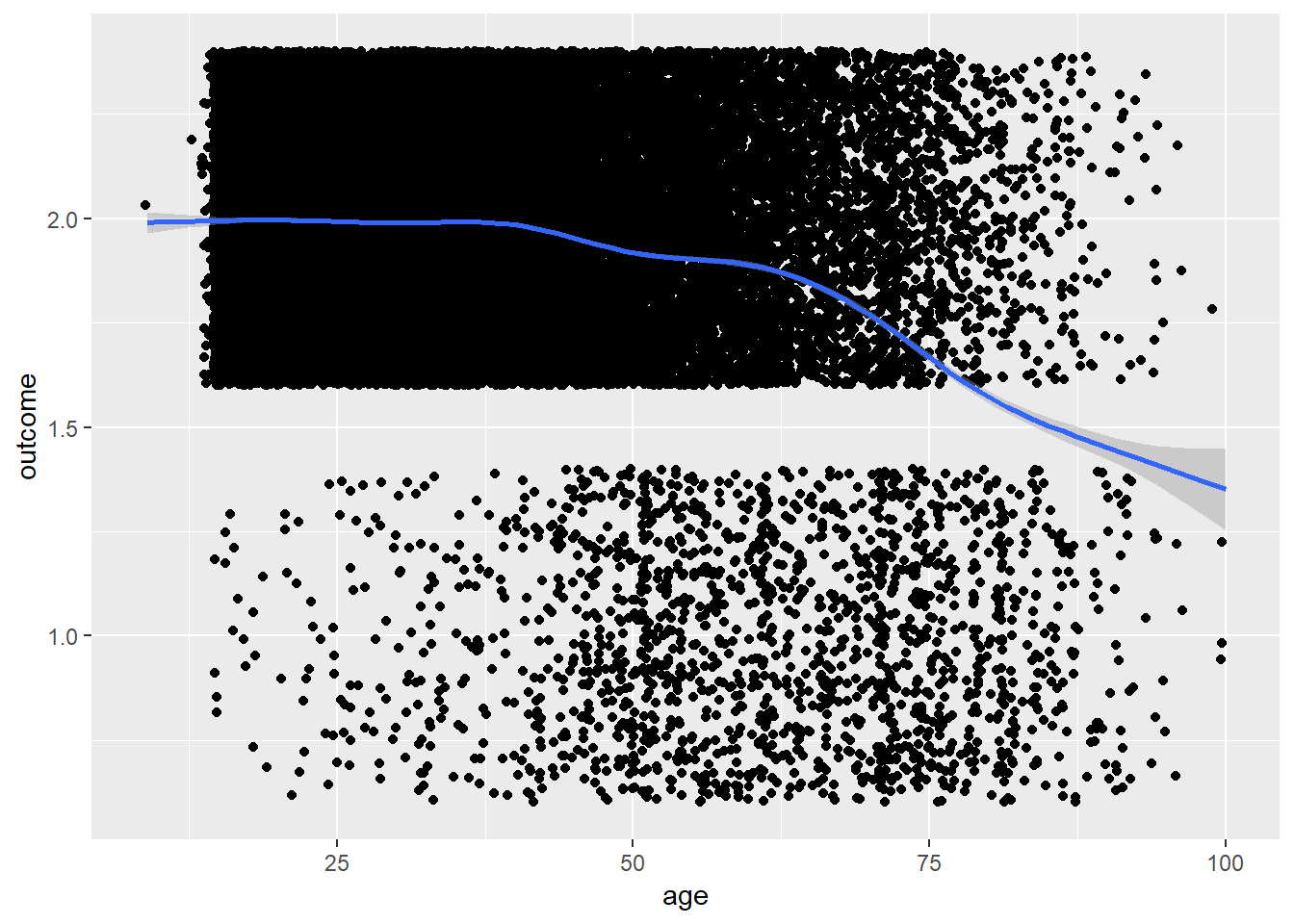
birthorder = birthorder %>%
mutate(outcome = Elementary_missed)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'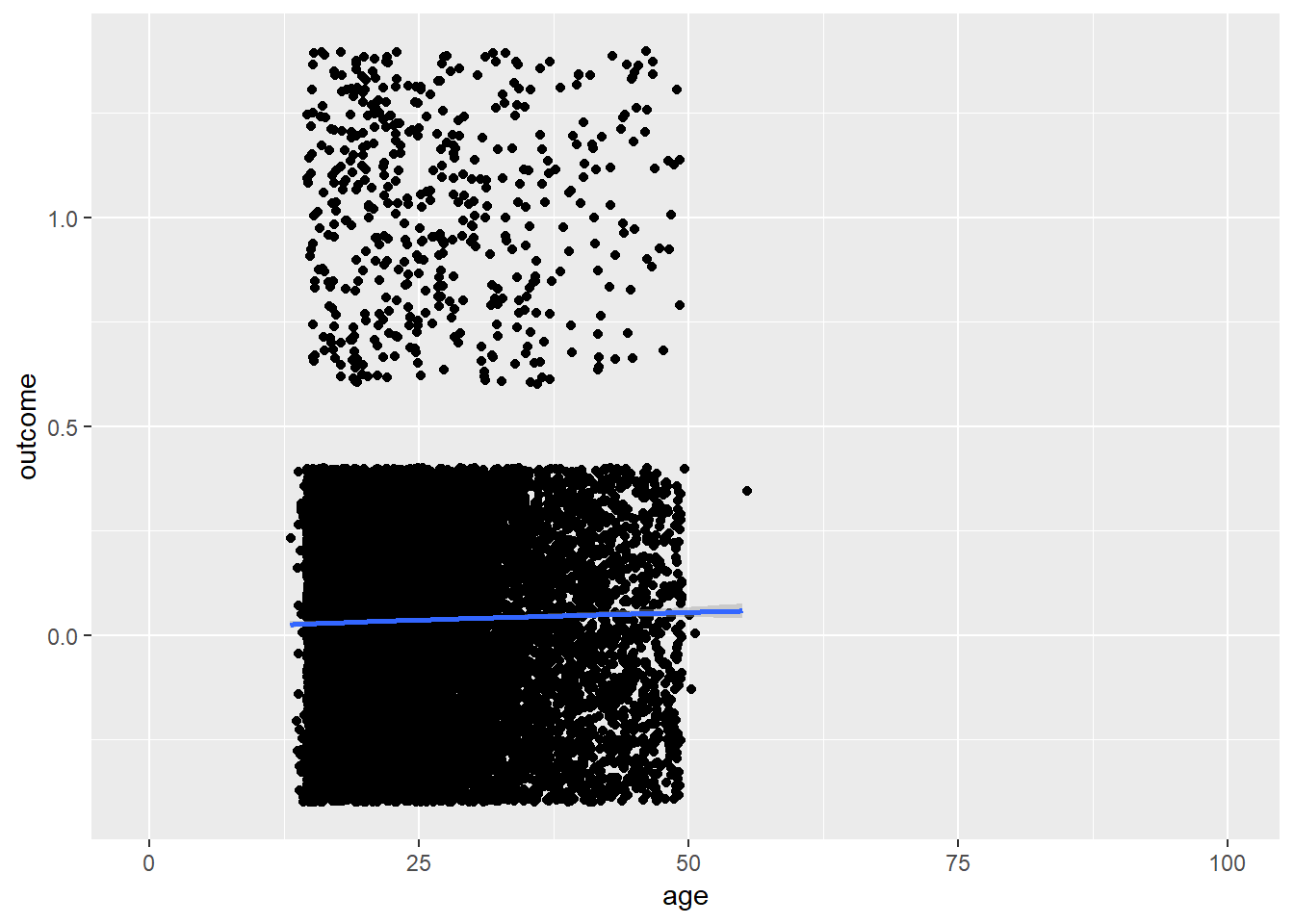
birthorder = birthorder %>%
mutate(outcome = Elementary_worked)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'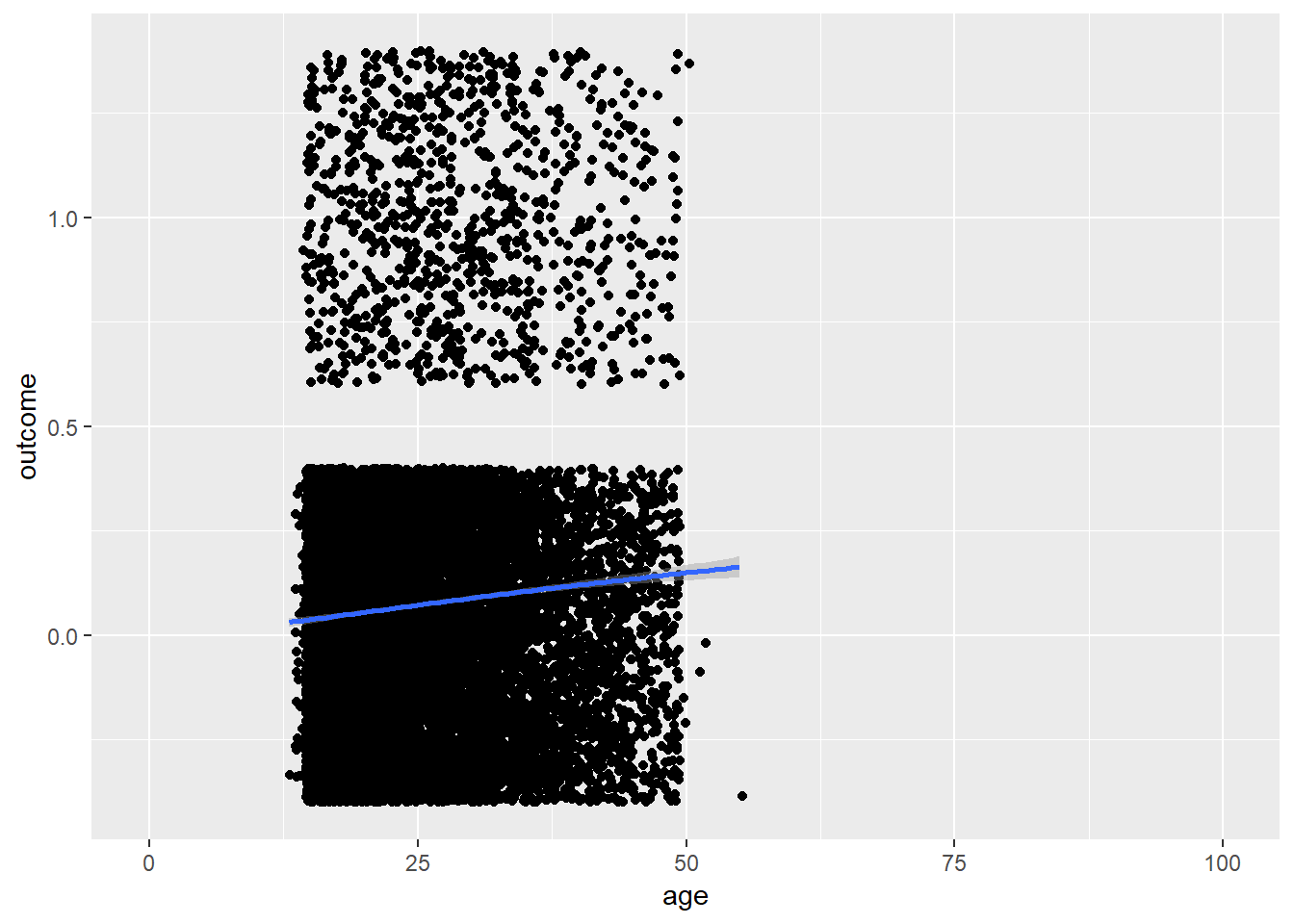
birthorder = birthorder %>%
mutate(outcome = wage_last_month_log)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'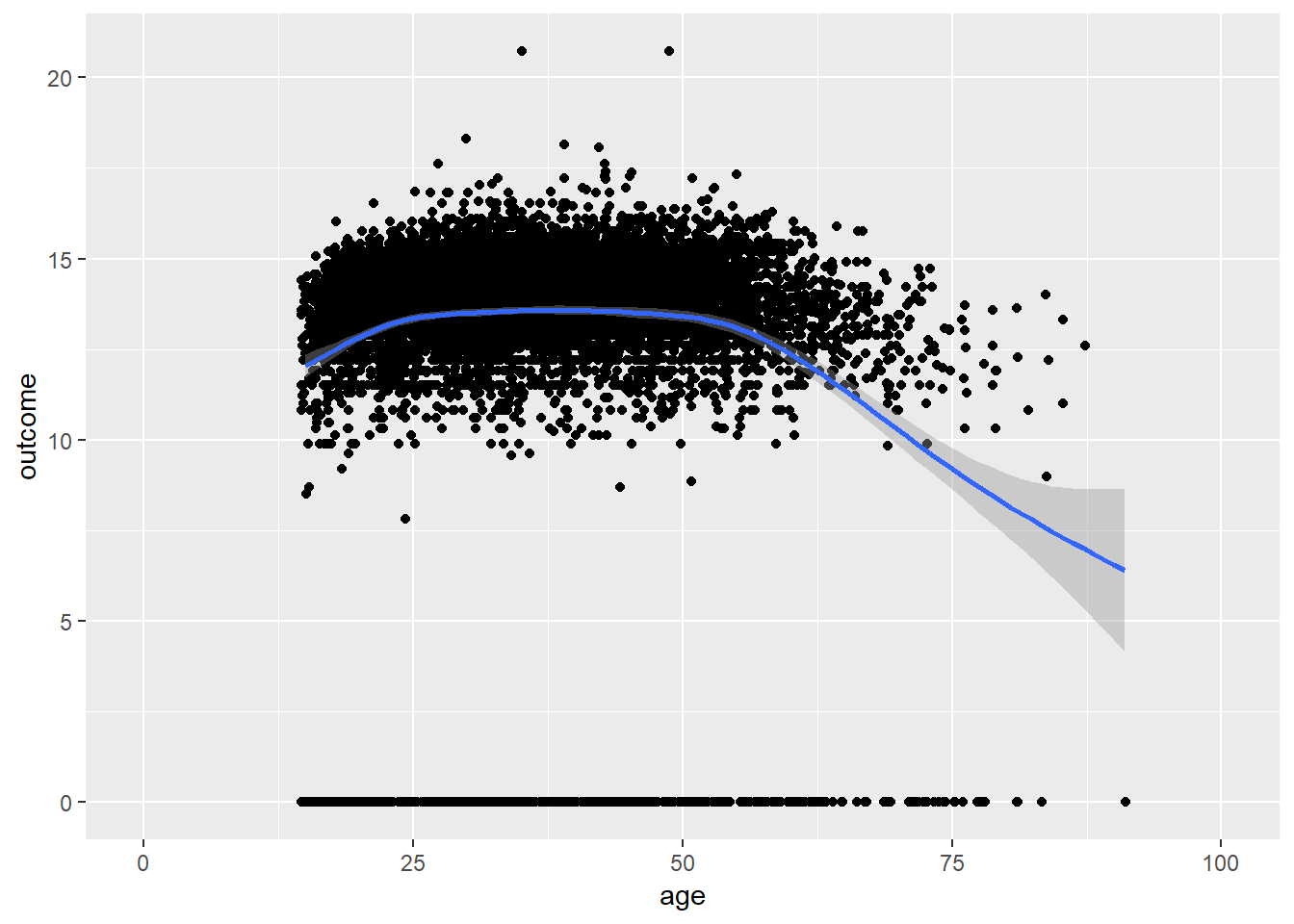
birthorder = birthorder %>%
mutate(outcome = wage_last_year_log)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'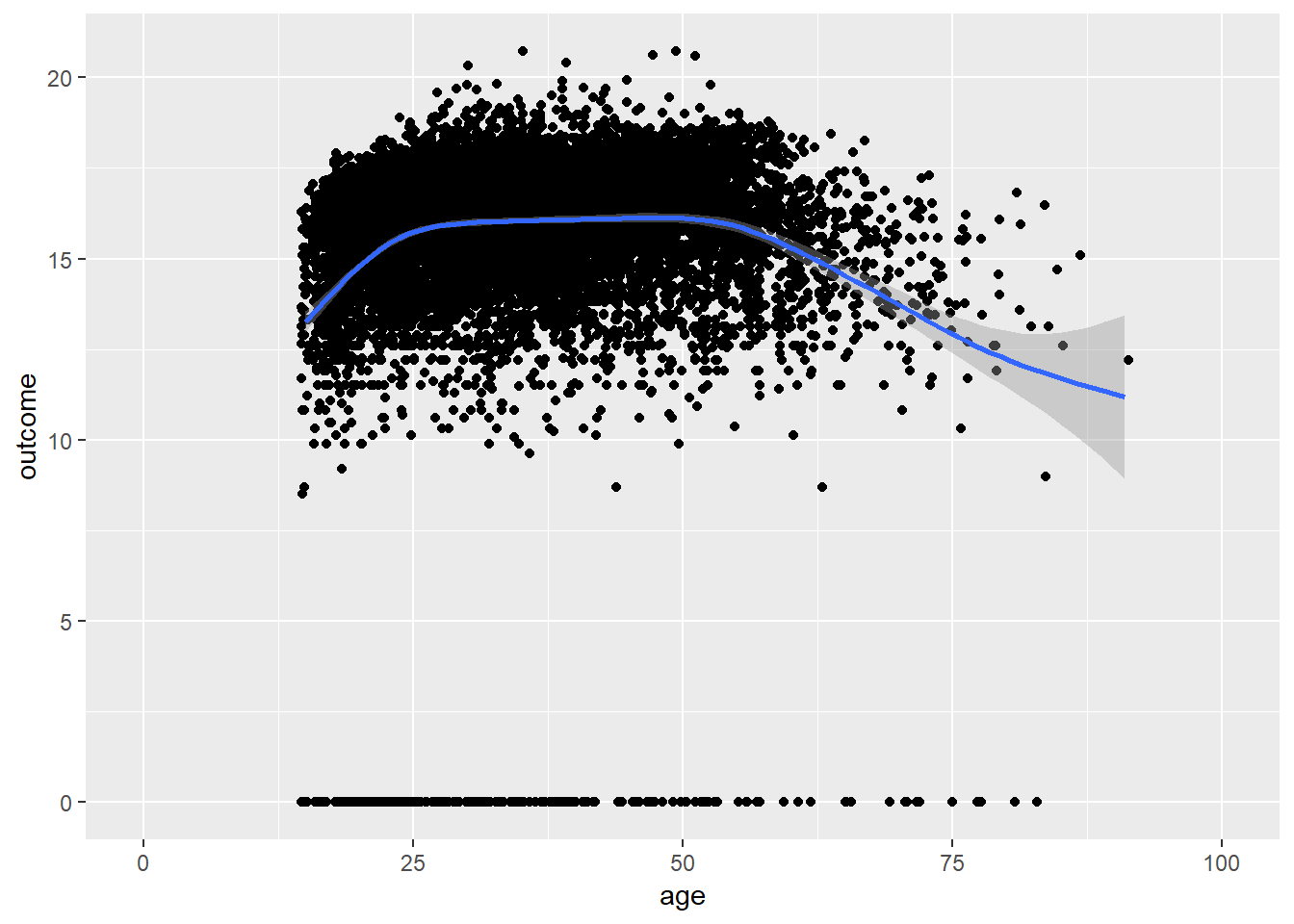
birthorder = birthorder %>%
mutate(outcome = Self_employed)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'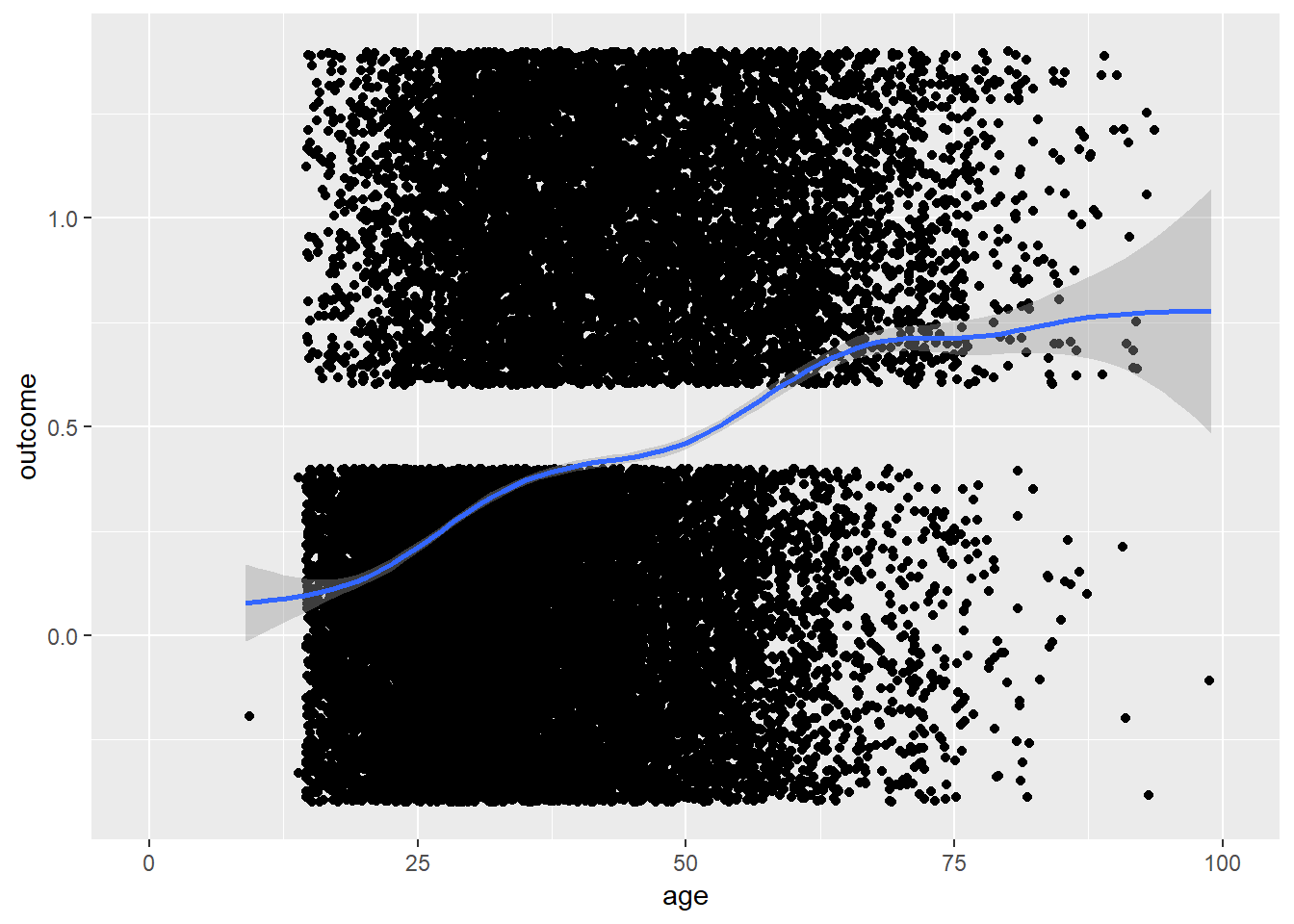
birthorder = birthorder %>%
mutate(outcome = ever_smoked)
plot_age(birthorder)## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'
LS0tCnRpdGxlOiAiM19zYW1wbGVfYW5hbHlzZXMiCmF1dGhvcjogIkxhdXJhIEJvdHpldCAmIFJ1YmVuIEFyc2xhbiIKb3V0cHV0OiBodG1sX2RvY3VtZW50CmVkaXRvcl9vcHRpb25zOiAKICBjaHVua19vdXRwdXRfdHlwZTogY29uc29sZQotLS0KIyAgPHNwYW4gc3R5bGU9ImNvbG9yOiNFNzhBQzMiPlNhbXBsZSBBbmFseXNlczwvc3Bhbj4gey50YWJzZXR9CgojIyBIZWxwZXIKYGBge3IgaGVscGVyfQpzb3VyY2UoIjBfaGVscGVycy5SIikKYGBgCgojIyBMb2FkIGRhdGEKYGBge3IgTG9hZCBkYXRhfQojIyMgSW1wb3J0IGFsbCBkYXRhIHdpdGgga25vd24gYmlydGhvcmRlcgpiaXJ0aG9yZGVyID0gcmVhZFJEUygiZGF0YS9hbGxkYXRhX2JpcnRob3JkZXIucmRzIikKYGBgCgojIyBPdmVydmlldyBSYXcgZGF0YQpgYGB7ciBPdmVydmlldyBSYXcgRGF0YX0KIyBjb2RlYm9vayhiaXJ0aG9yZGVyKQpgYGAKCiMjIERhdGEgd3JhbmdsaW5nCmBgYHtyIGRhdGEgd3JhbmdsaW5nfQojIyB3ZSBoYXZlIHRvIGV4Y2x1ZGUgcGVvcGxlIGluIHRoZSBjb250cm9sIGdyb3VwIHdobyBhcmUgcGFydCBvZiB0aGUgYmlydGhvcmRlciBncm91cApiaXJ0aG9yZGVyID0gYmlydGhvcmRlciAlPiUKICAjIG1hcmsgcGVvcGxlIHdobyBoYXZlIG1pc3NpbmcgYmlydGhvcmRlciBkYXRhCiAgbXV0YXRlKGNoZWNrX2JpcnRob3JkZXIgPSBpZmVsc2UoIWlzLm5hKGJpcnRob3JkZXJfZ2VuZXMpLCAxLCAwKSwKICAjIG1hcmsgcGVvcGxlIHdobyBoYXZlIG1pc3Npbmcgb3V0Y29tZXMKICAgICAgICAgY2hlY2tfb3V0Y29tZSA9IGlmZWxzZSghaXMubmEocmF2ZW5fMjAxNV9vbGQpLCAxLAogICAgICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKCFpcy5uYShtYXRoXzIwMTVfb2xkKSwgMSwKICAgICAgICAgICAgICAgICAgICAgICAgIGlmZWxzZSghaXMubmEocmF2ZW5fMjAxNV95b3VuZyksIDEsCiAgICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoIWlzLm5hKG1hdGhfMjAxNV95b3VuZyksIDEsCiAgICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoIWlzLm5hKHJhdmVuXzIwMDdfb2xkKSwgMSwKICAgICAgICAgICAgICAgICAgICAgICAgIGlmZWxzZSghaXMubmEobWF0aF8yMDA3X29sZCksIDEsCiAgICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoIWlzLm5hKHJhdmVuXzIwMDdfeW91bmcpLCAxLAogICAgICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKCFpcy5uYShtYXRoXzIwMDdfeW91bmcpLCAxLAogICAgICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKCFpcy5uYShhZGFwdGl2ZV9udW1iZXJpbmcpLCAxLAogICAgICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKCFpcy5uYSh3b3Jkc19yZW1lbWJlcmVkX2F2ZyksIDEsCiAgICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoIWlzLm5hKGNvdW50X2JhY2t3YXJkcyksIDEsCiAgICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoIWlzLm5hKGJpZzVfZXh0KSwgMSwgCiAgICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoIWlzLm5hKHJpc2tBKSwgMSwKICAgICAgICAgICAgICAgICAgICAgICAgIGlmZWxzZSghaXMubmEocmlza0IpLCAxLAogICAgICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKCFpcy5uYSh5ZWFyc19vZl9lZHVjYXRpb24pLCAxLAogICAgICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKCFpcy5uYShFbGVtZW50YXJ5X21pc3NlZCksIDEsCiAgICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoIWlzLm5hKEVsZW1lbnRhcnlfd29ya2VkKSwgMSwKICAgICAgICAgICAgICAgICAgICAgICAgIGlmZWxzZSghaXMubmEoYXR0ZW5kZWRfc2Nob29sKSwgMSwKICAgICAgICAgICAgICAgICAgICAgICAgIGlmZWxzZSghaXMubmEod2FnZV9sYXN0X21vbnRoX2xvZyksIDEsCiAgICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoIWlzLm5hKHdhZ2VfbGFzdF95ZWFyX2xvZyksIDEsCiAgICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoIWlzLm5hKFNlbGZfZW1wbG95ZWQpLCAxLAogICAgICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKCFpcy5uYShDYXRlZ29yeSksIDEsCiAgICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoIWlzLm5hKFNlY3RvciksIDEsCiAgICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoIWlzLm5hKGV2ZXJfc21va2VkKSwgMSwKICAgICAgICAgICAgICAgICAgICAgICAgIGlmZWxzZSghaXMubmEoc3RpbGxfc21va2luZyksIDEsCiAgICAgICAgICAgICAgICAgICAgICAgICAwKSkpKSkpKSkpKSkpKSkpKSkpKSkpKSkpKSwKICBncm91cF9iaXJ0aG9yZGVyID0gaWZlbHNlKGNoZWNrX2JpcnRob3JkZXIgPT0gMCwgImNvbnRyb2wiLCAidGVzdCIpLAogIGdyb3VwX291dGNvbWUgPSBpZmVsc2UoY2hlY2tfb3V0Y29tZSA9PSAwLCAiY29udHJvbCIsICJ0ZXN0IikpCmBgYAoKIyMgQmlydGggb3JkZXIgYW5kIHNpYnNoaXAgc2l6ZSB7LmFjdGl2ZSAudGFic2V0fQoKIyMjIEdlbmVzIGJpcnRob3JkZXIKYGBge3IgbWF0ZXJuYWwgYmlydGhvcmRlcn0KZGVzY3JpcHRpdmVzID0gYmlydGhvcmRlciAlPiUKICBncm91cF9ieShncm91cF9vdXRjb21lKSAlPiUKICBzdW1tYXJpc2UoaW5kaXZpZHVhbHMgPSBuKCksCiAgICAgICAgICAgIG1vdGhlcnMgPSBsZW5ndGgodW5pcXVlKG1vdGhlcl9waWRsaW5rKSksCiAgICAgICAgICAgIHNpYnNoaXBfc2l6ZV9tZWFuID0gbWVhbihzaWJsaW5nX2NvdW50X2dlbmVzLCBuYS5ybSA9IFQpLAogICAgICAgICAgICBzaWJzaGlwX3NpemVfY29uZmlkZW5jZV9sb3cgPSBtZWFuKHNpYmxpbmdfY291bnRfZ2VuZXMsIG5hLnJtID0gVCkgLSAKICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKHNpYmxpbmdfY291bnRfZ2VuZXMsIG5hLnJtID0gVCkvc3FydChuKCkpKSwKICAgICAgICAgICAgc2lic2hpcF9zaXplX2NvbmZpZGVuY2VfaGlnaCA9IG1lYW4oc2libGluZ19jb3VudF9nZW5lcywgbmEucm0gPSBUKSArIAogICAgICAgICAgICAgIChxdCguOTc1LCBuKCktMSkqc2Qoc2libGluZ19jb3VudF9nZW5lcywgbmEucm0gPSBUKS9zcXJ0KG4oKSkpLAogICAgICAgICAgICBzaWJzaGlwX3NpemVfbWluID0gbWluKHNpYmxpbmdfY291bnRfZ2VuZXMsIG5hLnJtID0gVFJVRSksCiAgICAgICAgICAgIHNpYnNoaXBfc2l6ZV9tYXggPSBtYXgoc2libGluZ19jb3VudF9nZW5lcywgbmEucm0gPSBUUlVFKSwKICAgICAgICAgICAgYmlydGhvcmRlcl9tZWFuID0gbWVhbihiaXJ0aG9yZGVyX2dlbmVzLCBuYS5ybSA9IFQpLAogICAgICAgICAgICBiaXJ0aG9yZGVyX3NpemVfY29uZmlkZW5jZV9sb3cgPSBtZWFuKGJpcnRob3JkZXJfZ2VuZXMsIG5hLnJtID0gVCkgLSAKICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKGJpcnRob3JkZXJfZ2VuZXMsIG5hLnJtID0gVCkvc3FydChuKCkpKSwKICAgICAgICAgICAgYmlydGhvcmRlcl9zaXplX2NvbmZpZGVuY2VfaGlnaCA9IG1lYW4oYmlydGhvcmRlcl9nZW5lcywgbmEucm0gPSBUKSArIAogICAgICAgICAgICAgIChxdCguOTc1LCBuKCktMSkqc2QoYmlydGhvcmRlcl9nZW5lcywgbmEucm0gPSBUKS9zcXJ0KG4oKSkpLAogICAgICAgICAgICBiaXJ0aG9yZGVyX3NpemVfbWluID0gbWluKGJpcnRob3JkZXJfZ2VuZXMsIG5hLnJtID0gVFJVRSksCiAgICAgICAgICAgIGJpcnRob3JkZXJfc2l6ZV9tYXggPSBtYXgoYmlydGhvcmRlcl9nZW5lcywgbmEucm0gPSBUUlVFKSwKICAgICAgICAgICAgbnVtYmVyX3NpYmxpbmdzX21lYW4gPSBtZWFuKHNpYmxpbmdfY291bnRfZ2VuZXMsIG5hLnJtID1UKSwKICAgICAgICAgICAgbnVtYmVyX3NpYmxpbmdzX2NvbmZpZGVuY2VfbG93ID0gbWVhbihzaWJsaW5nX2NvdW50X2dlbmVzLCBuYS5ybSA9IFQpIC0KICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKHNpYmxpbmdfY291bnRfZ2VuZXMsIG5hLnJtID0gVCkvc3FydChuKCkpKSwKICAgICAgICAgICAgbnVtYmVyX3NpYmxpbmdzX2NvbmZpZGVuY2VfaGlnaCA9IG1lYW4oc2libGluZ19jb3VudF9nZW5lcywgbmEucm0gPSBUKSArCiAgICAgICAgICAgICAgKHF0KC45NzUsIG4oKS0xKSpzZChzaWJsaW5nX2NvdW50X2dlbmVzLCBuYS5ybSA9IFQpL3NxcnQobigpKSkpCgpkZXNjcmlwdGl2ZXMKCmJpcnRob3JkZXIgJT4lIAogIHQudGVzdChzaWJsaW5nX2NvdW50X2dlbmVzIH4gZ3JvdXBfb3V0Y29tZSwgZGF0YSA9IC4sIHZhci5lcXVhbCA9IFQpCgpiaXJ0aG9yZGVyICU+JSAKICB0LnRlc3QoYmlydGhvcmRlcl9nZW5lcyB+IGdyb3VwX291dGNvbWUsIGRhdGEgPSAuLCB2YXIuZXF1YWwgPSBUKQoKYGBgCgoKIyMjIyBIb3cgbWFueSBzaWJsaW5ncyB3aXRoIGRhdGEgZG8gd2UgcmV0YWluIGluIGVhY2ggZmFtaWx5PwpgYGB7cn0KYmlydGhvcmRlciAlPiUgZmlsdGVyKCFpcy5uYShiaXJ0aG9yZGVyX2dlbmVzKSwgZ3JvdXBfb3V0Y29tZSA9PSAidGVzdCIpICU+JSBncm91cF9ieShtb3RoZXJfcGlkbGluaykgJT4lIHN1bW1hcmlzZSh3aXRoX2RhdGEgPSBuKCksIGFsbCA9IG1lYW4oc2libGluZ19jb3VudF9nZW5lcykpIC0+IGNvdW50cwpiaXJ0aG9yZGVyICU+JSBmaWx0ZXIoIWlzLm5hKGJpcnRob3JkZXJfZ2VuZXMpLCBncm91cF9vdXRjb21lID09ICJjb250cm9sIikgJT4lIGdyb3VwX2J5KG1vdGhlcl9waWRsaW5rKSAlPiUgc3VtbWFyaXNlKHdpdGhfZGF0YSA9IG4oKSwgYWxsID0gbWVhbihzaWJsaW5nX2NvdW50X2dlbmVzKSkgLT4gY291bnRzMQpgYGAKCkluIG91ciB0ZXN0IHNhbXBsZSBmYW1pbGllcyB3aXRoIGFuIGF2ZXJhZ2Ugc2l6ZSBvZiBgciBtZWFuKGNvdW50cyRhbGwsbmEucm09IFQpYCBzaWJsaW5ncywgd2UgcmV0YWluIGByIG1lYW4oY291bnRzJHdpdGhfZGF0YSlgLgoKSW4gdGhlIG9yaWdpbmFsIHNhbXBsZSBmYW1pbGllcyB3aXRoIGFuIGF2ZXJhZ2Ugc2l6ZSBvZiBgciBtZWFuKGNvdW50czEkYWxsLG5hLnJtPSBUKWAgc2libGluZ3MsIHdlIHJldGFpbiBgciBtZWFuKGNvdW50czEkd2l0aF9kYXRhKWAuCgpgYGB7cn0KZ2dwbG90KGNvdW50cywgYWVzKGFsbCwgd2l0aF9kYXRhKSkgKyBnZW9tX2ppdHRlcihhbHBoYSA9IDAuMSkgKyBnZW9tX3Ntb290aCgpICsgc2NhbGVfeF9jb250aW51b3VzKGJyZWFrcz0xOjE1KSArIHNjYWxlX3lfY29udGludW91cyhicmVha3M9MToxNSkKYGBgCgojIyMgTWF0ZXJuYWwgYmlydGhvcmRlcgpgYGB7ciBtYXRlcm5hbCBiaXJ0aG9yZGVyfQpkZXNjcmlwdGl2ZXMgPSBiaXJ0aG9yZGVyICU+JQogIGdyb3VwX2J5KGdyb3VwX291dGNvbWUpICU+JQogIHN1bW1hcmlzZShpbmRpdmlkdWFscyA9IG4oKSwKICAgICAgICAgICAgbW90aGVycyA9IGxlbmd0aCh1bmlxdWUobW90aGVyX3BpZGxpbmspKSwKICAgICAgICAgICAgc2lic2hpcF9zaXplX21lYW4gPSBtZWFuKHNpYmxpbmdfY291bnRfdXRlcnVzX2FsaXZlLCBuYS5ybSA9IFQpLAogICAgICAgICAgICBzaWJzaGlwX3NpemVfY29uZmlkZW5jZV9sb3cgPSBtZWFuKHNpYmxpbmdfY291bnRfdXRlcnVzX2FsaXZlLCBuYS5ybSA9IFQpIC0gCiAgICAgICAgICAgICAgKHF0KC45NzUsIG4oKS0xKSpzZChzaWJsaW5nX2NvdW50X3V0ZXJ1c19hbGl2ZSwgbmEucm0gPSBUKS9zcXJ0KG4oKSkpLAogICAgICAgICAgICBzaWJzaGlwX3NpemVfY29uZmlkZW5jZV9oaWdoID0gbWVhbihzaWJsaW5nX2NvdW50X3V0ZXJ1c19hbGl2ZSwgbmEucm0gPSBUKSArIAogICAgICAgICAgICAgIChxdCguOTc1LCBuKCktMSkqc2Qoc2libGluZ19jb3VudF91dGVydXNfYWxpdmUsIG5hLnJtID0gVCkvc3FydChuKCkpKSwKICAgICAgICAgICAgc2lic2hpcF9zaXplX21pbiA9IG1pbihzaWJsaW5nX2NvdW50X3V0ZXJ1c19hbGl2ZSwgbmEucm0gPSBUUlVFKSwKICAgICAgICAgICAgc2lic2hpcF9zaXplX21heCA9IG1heChzaWJsaW5nX2NvdW50X3V0ZXJ1c19hbGl2ZSwgbmEucm0gPSBUUlVFKSwKICAgICAgICAgICAgYmlydGhvcmRlcl9tZWFuID0gbWVhbihiaXJ0aG9yZGVyX3V0ZXJ1c19hbGl2ZSwgbmEucm0gPSBUKSwKICAgICAgICAgICAgYmlydGhvcmRlcl9zaXplX2NvbmZpZGVuY2VfbG93ID0gbWVhbihiaXJ0aG9yZGVyX3V0ZXJ1c19hbGl2ZSwgbmEucm0gPSBUKSAtIAogICAgICAgICAgICAgIChxdCguOTc1LCBuKCktMSkqc2QoYmlydGhvcmRlcl91dGVydXNfYWxpdmUsIG5hLnJtID0gVCkvc3FydChuKCkpKSwKICAgICAgICAgICAgYmlydGhvcmRlcl9zaXplX2NvbmZpZGVuY2VfaGlnaCA9IG1lYW4oYmlydGhvcmRlcl91dGVydXNfYWxpdmUsIG5hLnJtID0gVCkgKyAKICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKGJpcnRob3JkZXJfdXRlcnVzX2FsaXZlLCBuYS5ybSA9IFQpL3NxcnQobigpKSksCiAgICAgICAgICAgIGJpcnRob3JkZXJfc2l6ZV9taW4gPSBtaW4oYmlydGhvcmRlcl91dGVydXNfYWxpdmUsIG5hLnJtID0gVFJVRSksCiAgICAgICAgICAgIGJpcnRob3JkZXJfc2l6ZV9tYXggPSBtYXgoYmlydGhvcmRlcl91dGVydXNfYWxpdmUsIG5hLnJtID0gVFJVRSksCiAgICAgICAgICAgIG51bWJlcl9zaWJsaW5nc19tZWFuID0gbWVhbihzaWJsaW5nX2NvdW50X3V0ZXJ1c19hbGl2ZSwgbmEucm0gPVQpLAogICAgICAgICAgICBudW1iZXJfc2libGluZ3NfY29uZmlkZW5jZV9sb3cgPSBtZWFuKHNpYmxpbmdfY291bnRfdXRlcnVzX2FsaXZlLCBuYS5ybSA9IFQpIC0KICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKHNpYmxpbmdfY291bnRfdXRlcnVzX2FsaXZlLCBuYS5ybSA9IFQpL3NxcnQobigpKSksCiAgICAgICAgICAgIG51bWJlcl9zaWJsaW5nc19jb25maWRlbmNlX2hpZ2ggPSBtZWFuKHNpYmxpbmdfY291bnRfdXRlcnVzX2FsaXZlLCBuYS5ybSA9IFQpICsKICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKHNpYmxpbmdfY291bnRfdXRlcnVzX2FsaXZlLCBuYS5ybSA9IFQpL3NxcnQobigpKSkpCgpkZXNjcmlwdGl2ZXMKCmJpcnRob3JkZXIgJT4lIAogIHQudGVzdChzaWJsaW5nX2NvdW50X3V0ZXJ1c19hbGl2ZSB+IGdyb3VwX291dGNvbWUsIGRhdGEgPSAuLCB2YXIuZXF1YWwgPSBUKQoKYmlydGhvcmRlciAlPiUgCiAgdC50ZXN0KGJpcnRob3JkZXJfdXRlcnVzX2FsaXZlIH4gZ3JvdXBfb3V0Y29tZSwgZGF0YSA9IC4sIHZhci5lcXVhbCA9IFQpCgpgYGAKCgojIyMjIEhvdyBtYW55IHNpYmxpbmdzIHdpdGggZGF0YSBkbyB3ZSByZXRhaW4gaW4gZWFjaCBmYW1pbHk/CmBgYHtyfQoKYmlydGhvcmRlciAlPiUgZmlsdGVyKCFpcy5uYShiaXJ0aG9yZGVyX3V0ZXJ1c19hbGl2ZSksIGdyb3VwX291dGNvbWUgPT0gMSkgJT4lIGdyb3VwX2J5KG1vdGhlcl9waWRsaW5rKSAlPiUgc3VtbWFyaXNlKHdpdGhfZGF0YSA9IG4oKSwgYWxsID0gbWVhbihzaWJsaW5nX2NvdW50X3V0ZXJ1c19hbGl2ZSkpIC0+IGNvdW50cwpiaXJ0aG9yZGVyICU+JSBmaWx0ZXIoIWlzLm5hKGJpcnRob3JkZXJfdXRlcnVzX2FsaXZlKSwgZ3JvdXBfb3V0Y29tZSA9PSAwKSAlPiUgZ3JvdXBfYnkobW90aGVyX3BpZGxpbmspICU+JSBzdW1tYXJpc2Uod2l0aF9kYXRhID0gbigpLCBhbGwgPSBtZWFuKHNpYmxpbmdfY291bnRfdXRlcnVzX2FsaXZlKSkgLT4gY291bnRzMQpgYGAKCkluIG91ciB0ZXN0IHNhbXBsZSBmYW1pbGllcyB3aXRoIGFuIGF2ZXJhZ2Ugc2l6ZSBvZiBgciBtZWFuKGNvdW50cyRhbGwsbmEucm09IFQpYCBzaWJsaW5ncywgd2UgcmV0YWluIGByIG1lYW4oY291bnRzJHdpdGhfZGF0YSlgLgoKSW4gdGhlIG9yaWdpbmFsIHNhbXBsZSBmYW1pbGllcyB3aXRoIGFuIGF2ZXJhZ2Ugc2l6ZSBvZiBgciBtZWFuKGNvdW50czEkYWxsLG5hLnJtPSBUKWAgc2libGluZ3MsIHdlIHJldGFpbiBgciBtZWFuKGNvdW50czEkd2l0aF9kYXRhKWAuCgpgYGB7cn0KZ2dwbG90KGNvdW50cywgYWVzKGFsbCwgd2l0aF9kYXRhKSkgKyBnZW9tX2ppdHRlcihhbHBoYSA9IDAuMSkgKyBnZW9tX3Ntb290aCgpICsgc2NhbGVfeF9jb250aW51b3VzKGJyZWFrcz0xOjE1KSArIHNjYWxlX3lfY29udGludW91cyhicmVha3M9MToxNSkKYGBgCiMjIyBNYXRlcm5hbCBwcmVnbmFuY3kgYmlydGhvcmRlcgpgYGB7ciBtYXRlcm5hbCBiaXJ0aG9yZGVyfQpkZXNjcmlwdGl2ZXMgPSBiaXJ0aG9yZGVyICU+JQogIGdyb3VwX2J5KGdyb3VwX291dGNvbWUpICU+JQogIHN1bW1hcmlzZShpbmRpdmlkdWFscyA9IG4oKSwKICAgICAgICAgICAgbW90aGVycyA9IGxlbmd0aCh1bmlxdWUobW90aGVyX3BpZGxpbmspKSwKICAgICAgICAgICAgc2lic2hpcF9zaXplX21lYW4gPSBtZWFuKHNpYmxpbmdfY291bnRfdXRlcnVzX3ByZWcsIG5hLnJtID0gVCksCiAgICAgICAgICAgIHNpYnNoaXBfc2l6ZV9jb25maWRlbmNlX2xvdyA9IG1lYW4oc2libGluZ19jb3VudF91dGVydXNfcHJlZywgbmEucm0gPSBUKSAtIAogICAgICAgICAgICAgIChxdCguOTc1LCBuKCktMSkqc2Qoc2libGluZ19jb3VudF91dGVydXNfcHJlZywgbmEucm0gPSBUKS9zcXJ0KG4oKSkpLAogICAgICAgICAgICBzaWJzaGlwX3NpemVfY29uZmlkZW5jZV9oaWdoID0gbWVhbihzaWJsaW5nX2NvdW50X3V0ZXJ1c19wcmVnLCBuYS5ybSA9IFQpICsgCiAgICAgICAgICAgICAgKHF0KC45NzUsIG4oKS0xKSpzZChzaWJsaW5nX2NvdW50X3V0ZXJ1c19wcmVnLCBuYS5ybSA9IFQpL3NxcnQobigpKSksCiAgICAgICAgICAgIHNpYnNoaXBfc2l6ZV9taW4gPSBtaW4oc2libGluZ19jb3VudF91dGVydXNfcHJlZywgbmEucm0gPSBUUlVFKSwKICAgICAgICAgICAgc2lic2hpcF9zaXplX21heCA9IG1heChzaWJsaW5nX2NvdW50X3V0ZXJ1c19wcmVnLCBuYS5ybSA9IFRSVUUpLAogICAgICAgICAgICBiaXJ0aG9yZGVyX21lYW4gPSBtZWFuKGJpcnRob3JkZXJfdXRlcnVzX3ByZWcsIG5hLnJtID0gVCksCiAgICAgICAgICAgIGJpcnRob3JkZXJfc2l6ZV9jb25maWRlbmNlX2xvdyA9IG1lYW4oYmlydGhvcmRlcl91dGVydXNfcHJlZywgbmEucm0gPSBUKSAtIAogICAgICAgICAgICAgIChxdCguOTc1LCBuKCktMSkqc2QoYmlydGhvcmRlcl91dGVydXNfcHJlZywgbmEucm0gPSBUKS9zcXJ0KG4oKSkpLAogICAgICAgICAgICBiaXJ0aG9yZGVyX3NpemVfY29uZmlkZW5jZV9oaWdoID0gbWVhbihiaXJ0aG9yZGVyX3V0ZXJ1c19wcmVnLCBuYS5ybSA9IFQpICsgCiAgICAgICAgICAgICAgKHF0KC45NzUsIG4oKS0xKSpzZChiaXJ0aG9yZGVyX3V0ZXJ1c19wcmVnLCBuYS5ybSA9IFQpL3NxcnQobigpKSksCiAgICAgICAgICAgIGJpcnRob3JkZXJfc2l6ZV9taW4gPSBtaW4oYmlydGhvcmRlcl91dGVydXNfcHJlZywgbmEucm0gPSBUUlVFKSwKICAgICAgICAgICAgYmlydGhvcmRlcl9zaXplX21heCA9IG1heChiaXJ0aG9yZGVyX3V0ZXJ1c19wcmVnLCBuYS5ybSA9IFRSVUUpLAogICAgICAgICAgICBudW1iZXJfc2libGluZ3NfbWVhbiA9IG1lYW4oc2libGluZ19jb3VudF91dGVydXNfcHJlZywgbmEucm0gPVQpLAogICAgICAgICAgICBudW1iZXJfc2libGluZ3NfY29uZmlkZW5jZV9sb3cgPSBtZWFuKHNpYmxpbmdfY291bnRfdXRlcnVzX3ByZWcsIG5hLnJtID0gVCkgLQogICAgICAgICAgICAgIChxdCguOTc1LCBuKCktMSkqc2Qoc2libGluZ19jb3VudF91dGVydXNfcHJlZywgbmEucm0gPSBUKS9zcXJ0KG4oKSkpLAogICAgICAgICAgICBudW1iZXJfc2libGluZ3NfY29uZmlkZW5jZV9oaWdoID0gbWVhbihzaWJsaW5nX2NvdW50X3V0ZXJ1c19wcmVnLCBuYS5ybSA9IFQpICsKICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKHNpYmxpbmdfY291bnRfdXRlcnVzX3ByZWcsIG5hLnJtID0gVCkvc3FydChuKCkpKSkKCmRlc2NyaXB0aXZlcwoKYmlydGhvcmRlciAlPiUgCiAgdC50ZXN0KHNpYmxpbmdfY291bnRfdXRlcnVzX3ByZWcgfiBncm91cF9vdXRjb21lLCBkYXRhID0gLiwgdmFyLmVxdWFsID0gVCkKCmJpcnRob3JkZXIgJT4lIAogIHQudGVzdChiaXJ0aG9yZGVyX3V0ZXJ1c19wcmVnIH4gZ3JvdXBfb3V0Y29tZSwgZGF0YSA9IC4sIHZhci5lcXVhbCA9IFQpCgpgYGAKCgojIyMjIEhvdyBtYW55IHNpYmxpbmdzIHdpdGggZGF0YSBkbyB3ZSByZXRhaW4gaW4gZWFjaCBmYW1pbHk/CmBgYHtyfQpiaXJ0aG9yZGVyICU+JSBmaWx0ZXIoIWlzLm5hKGJpcnRob3JkZXJfdXRlcnVzX3ByZWcpLCBncm91cF9vdXRjb21lID09IDEpICU+JSBncm91cF9ieShtb3RoZXJfcGlkbGluaykgJT4lIHN1bW1hcmlzZSh3aXRoX2RhdGEgPSBuKCksIGFsbCA9IG1lYW4oc2libGluZ19jb3VudF91dGVydXNfcHJlZykpIC0+IGNvdW50cwpiaXJ0aG9yZGVyICU+JSBmaWx0ZXIoIWlzLm5hKGJpcnRob3JkZXJfdXRlcnVzX3ByZWcpLCBncm91cF9vdXRjb21lID09IDApICU+JSBncm91cF9ieShtb3RoZXJfcGlkbGluaykgJT4lIHN1bW1hcmlzZSh3aXRoX2RhdGEgPSBuKCksIGFsbCA9IG1lYW4oc2libGluZ19jb3VudF91dGVydXNfcHJlZykpIC0+IGNvdW50czEKYGBgCgpJbiBvdXIgdGVzdCBzYW1wbGUgZmFtaWxpZXMgd2l0aCBhbiBhdmVyYWdlIHNpemUgb2YgYHIgbWVhbihjb3VudHMkYWxsLG5hLnJtPSBUKWAgc2libGluZ3MsIHdlIHJldGFpbiBgciBtZWFuKGNvdW50cyR3aXRoX2RhdGEpYC4KCkluIHRoZSBvcmlnaW5hbCBzYW1wbGUgZmFtaWxpZXMgd2l0aCBhbiBhdmVyYWdlIHNpemUgb2YgYHIgbWVhbihjb3VudHMxJGFsbCxuYS5ybT0gVClgIHNpYmxpbmdzLCB3ZSByZXRhaW4gYHIgbWVhbihjb3VudHMxJHdpdGhfZGF0YSlgLgoKYGBge3J9CmdncGxvdChjb3VudHMsIGFlcyhhbGwsIHdpdGhfZGF0YSkpICsgZ2VvbV9qaXR0ZXIoYWxwaGEgPSAwLjEpICsgZ2VvbV9zbW9vdGgoKSArIHNjYWxlX3hfY29udGludW91cyhicmVha3M9MToxNSkgKyBzY2FsZV95X2NvbnRpbnVvdXMoYnJlYWtzPTE6MTUpCmBgYAoKCiMjIE91dGNvbWUgbWVhc3VyZW1lbnRzIGFuZCBjb3ZhcmlhdGVzIHsudGFic2V0fQojIyMgQ29udHJvbCBncm91cApgYGB7ciBkYXRhIGNvbXBhcmlzb259CiMjIERlc2NyaXB0aXZlcwpkZXNjcmlwdGl2ZXMgPSBiaXJ0aG9yZGVyICU+JQogIGdyb3VwX2J5KGdyb3VwX2JpcnRob3JkZXIpICU+JQogICAgc3VtbWFyaXNlKG4gPSBuKCksCiAgICAgICAgICAgICAgYWdlX21lYW4gPSBtZWFuKGFnZSwgbmEucm09VFJVRSksCiAgICAgICAgICAgICAgYWdlX2NvbmZpZGVuY2VfbG93ID0gbWVhbihhZ2UsIG5hLnJtID0gVCkgLSAKICAgICAgICAgICAgICAgIChxdCguOTc1LCBuKCktMSkqc2QoYWdlLCBuYS5ybSA9IFQpL3NxcnQobigpKSksCiAgICAgICAgICAgICAgYWdlX2NvbmZpZGVuY2VfaGlnaCA9IG1lYW4oYWdlLCBuYS5ybSA9IFQpICsgCiAgICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKGFnZSwgbmEucm0gPSBUKS9zcXJ0KG4oKSkpLAogICAgICAgICAgICAgIGFnZV9taW4gPSBtaW4oYWdlLCBuYS5ybSA9IFRSVUUpLAogICAgICAgICAgICAgIGFnZV9tYXggPSBtYXgoYWdlLCBuYS5ybSA9IFRSVUUpLAogICAgICAgICAgICAgIGdlbmRlciA9IG1lYW4obWFsZSwgbmEucm09VFJVRSkpCmRlc2NyaXB0aXZlcwoKCgojIyBUdGVzdAp0aWR5KHQudGVzdChiaXJ0aG9yZGVyJGFnZSB+IGJpcnRob3JkZXIkZ3JvdXBfYmlydGhvcmRlciwgdmFyLmVxdWFsID0gVCkpCgpjb2hlbi5kKGJpcnRob3JkZXIkYWdlLCBhcy5mYWN0b3IoYmlydGhvcmRlciRncm91cF9iaXJ0aG9yZGVyKSwgbmEucm0gPSBUKQoKZ2VuZGVyID0gYmlydGhvcmRlciAlPiUKICBncm91cF9ieShncm91cF9iaXJ0aG9yZGVyKSAlPiUKICAgIHN1bW1hcmlzZShnZW5kZXIgPSBzdW0obWFsZSA9PSAxLCBuYS5ybT1UKSwKICAgICAgICAgICAgICBnZW5kZXIyID0gc3VtKG1hbGUgPT0wLG5hLnJtPVQpKSAlPiUKICBzZWxlY3QoZ2VuZGVyLCBnZW5kZXIyKQpwcm9wLnRhYmxlKGdlbmRlcikKdGlkeShjaGlzcS50ZXN0KGdlbmRlcikpCgojIyBSYXRpbmdzCnJhdGluZ3MgPSBiaXJ0aG9yZGVyICU+JQogIGdyb3VwX2J5KGdyb3VwX2JpcnRob3JkZXIpICU+JQogIHN1bW1hcmlzZShnX2ZhY3Rvcl9tZWFuID0gbWVhbihnX2ZhY3Rvcl8yMDE1X29sZCwgbmEucm09VCksCiAgICAgICAgICAgIGdfZmFjdG9yX2NvbmZpZGVuY2VfbG93PSBtZWFuKGdfZmFjdG9yXzIwMTVfb2xkLCBuYS5ybSA9IFQpIC0gCiAgICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKGdfZmFjdG9yXzIwMTVfb2xkLCBuYS5ybSA9IFQpL3NxcnQobigpKSksCiAgICAgICAgICAgIGdfZmFjdG9yX2NvbmZpZGVuY2VfaGlnaCA9IG1lYW4oZ19mYWN0b3JfMjAxNV9vbGQsIG5hLnJtID0gVCkgKwogICAgICAgICAgICAgICAgKHF0KC45NzUsIG4oKS0xKSpzZChnX2ZhY3Rvcl8yMDE1X29sZCwgbmEucm0gPSBUKS9zcXJ0KG4oKSkpLAogICAgICAgICAgICBiaWc1X2V4dF9tZWFuID0gbWVhbihiaWc1X2V4dCwgbmEucm09VCksCiAgICAgICAgICAgIGJpZzVfZXh0X2NvbmZpZGVuY2VfbG93PSBtZWFuKGJpZzVfZXh0LCBuYS5ybSA9IFQpIC0gCiAgICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKGJpZzVfZXh0LCBuYS5ybSA9IFQpL3NxcnQobigpKSksCiAgICAgICAgICAgIGJpZzVfZXh0X2NvbmZpZGVuY2VfaGlnaCA9IG1lYW4oYmlnNV9leHQsIG5hLnJtID0gVCkgKwogICAgICAgICAgICAgICAgKHF0KC45NzUsIG4oKS0xKSpzZChiaWc1X2V4dCwgbmEucm0gPSBUKS9zcXJ0KG4oKSkpLAogICAgICAgICAgICBiaWc1X25ldV9tZWFuID0gbWVhbihiaWc1X25ldSwgbmEucm09VCksCiAgICAgICAgICAgIGJpZzVfbmV1X2NvbmZpZGVuY2VfbG93PSBtZWFuKGJpZzVfbmV1LCBuYS5ybSA9IFQpIC0gCiAgICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKGJpZzVfbmV1LCBuYS5ybSA9IFQpL3NxcnQobigpKSksCiAgICAgICAgICAgIGJpZzVfbmV1X2NvbmZpZGVuY2VfaGlnaCA9IG1lYW4oYmlnNV9uZXUsIG5hLnJtID0gVCkgKwogICAgICAgICAgICAgICAgKHF0KC45NzUsIG4oKS0xKSpzZChiaWc1X25ldSwgbmEucm0gPSBUKS9zcXJ0KG4oKSkpLAogICAgICAgICAgICBiaWc1X2Nvbl9tZWFuID0gbWVhbihiaWc1X2NvbiwgbmEucm09VCksCiAgICAgICAgICAgIGJpZzVfY29uX2NvbmZpZGVuY2VfbG93PSBtZWFuKGJpZzVfY29uLCBuYS5ybSA9IFQpIC0gCiAgICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKGJpZzVfY29uLCBuYS5ybSA9IFQpL3NxcnQobigpKSksCiAgICAgICAgICAgIGJpZzVfY29uX2NvbmZpZGVuY2VfaGlnaCA9IG1lYW4oYmlnNV9jb24sIG5hLnJtID0gVCkgKwogICAgICAgICAgICAgICAgKHF0KC45NzUsIG4oKS0xKSpzZChiaWc1X2NvbiwgbmEucm0gPSBUKS9zcXJ0KG4oKSkpLAogICAgICAgICAgICBiaWc1X2FncmVlX21lYW4gPSBtZWFuKGJpZzVfYWdyZWUsIG5hLnJtPVQpLAogICAgICAgICAgICBiaWc1X2FncmVlX2NvbmZpZGVuY2VfbG93PSBtZWFuKGJpZzVfYWdyZWUsIG5hLnJtID0gVCkgLSAKICAgICAgICAgICAgICAgIChxdCguOTc1LCBuKCktMSkqc2QoYmlnNV9hZ3JlZSwgbmEucm0gPSBUKS9zcXJ0KG4oKSkpLAogICAgICAgICAgICBiaWc1X2FncmVlX2NvbmZpZGVuY2VfaGlnaCA9IG1lYW4oYmlnNV9hZ3JlZSwgbmEucm0gPSBUKSArCiAgICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKGJpZzVfYWdyZWUsIG5hLnJtID0gVCkvc3FydChuKCkpKSwKICAgICAgICAgICAgYmlnNV9vcGVuX21lYW4gPSBtZWFuKGJpZzVfb3BlbiwgbmEucm09VCksCiAgICAgICAgICAgIGJpZzVfb3Blbl9jb25maWRlbmNlX2xvdz0gbWVhbihiaWc1X29wZW4sIG5hLnJtID0gVCkgLSAKICAgICAgICAgICAgICAgIChxdCguOTc1LCBuKCktMSkqc2QoYmlnNV9vcGVuLCBuYS5ybSA9IFQpL3NxcnQobigpKSksCiAgICAgICAgICAgIGJpZzVfb3Blbl9jb25maWRlbmNlX2hpZ2ggPSBtZWFuKGJpZzVfb3BlbiwgbmEucm0gPSBUKSArCiAgICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKGJpZzVfb3BlbiwgbmEucm0gPSBUKS9zcXJ0KG4oKSkpLAogICAgICAgICAgICByaXNrQV9tZWFuID0gbWVhbihyaXNrQSwgbmEucm09VCksCiAgICAgICAgICAgIHJpc2tBX2NvbmZpZGVuY2VfbG93PSBtZWFuKHJpc2tBLCBuYS5ybSA9IFQpIC0gCiAgICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKHJpc2tBLCBuYS5ybSA9IFQpL3NxcnQobigpKSksCiAgICAgICAgICAgIHJpc2tBX2NvbmZpZGVuY2VfaGlnaCA9IG1lYW4ocmlza0EsIG5hLnJtID0gVCkgKwogICAgICAgICAgICAgICAgKHF0KC45NzUsIG4oKS0xKSpzZChyaXNrQSwgbmEucm0gPSBUKS9zcXJ0KG4oKSkpLAogICAgICAgICAgICByaXNrQl9tZWFuID0gbWVhbihyaXNrQiwgbmEucm09VCksCiAgICAgICAgICAgIHJpc2tCX2NvbmZpZGVuY2VfbG93PSBtZWFuKHJpc2tCLCBuYS5ybSA9IFQpIC0gCiAgICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKHJpc2tCLCBuYS5ybSA9IFQpL3NxcnQobigpKSksCiAgICAgICAgICAgIHJpc2tCX2NvbmZpZGVuY2VfaGlnaCA9IG1lYW4ocmlza0IsIG5hLnJtID0gVCkgKwogICAgICAgICAgICAgICAgKHF0KC45NzUsIG4oKS0xKSpzZChyaXNrQiwgbmEucm0gPSBUKS9zcXJ0KG4oKSkpKQpyYXRpbmdzCgp0aWR5KHQudGVzdChiaXJ0aG9yZGVyJGdfZmFjdG9yXzIwMTVfb2xkIH4gYmlydGhvcmRlciRncm91cF9iaXJ0aG9yZGVyLCB2YXIuZXF1YWwgPSBUKSkKY29oZW4uZChiaXJ0aG9yZGVyJGdfZmFjdG9yXzIwMTVfb2xkLCBhcy5mYWN0b3IoYmlydGhvcmRlciRncm91cF9iaXJ0aG9yZGVyKSwgbmEucm0gPSBUKQoKCnRpZHkodC50ZXN0KGJpcnRob3JkZXIkeWVhcnNfb2ZfZWR1Y2F0aW9uIH4gYmlydGhvcmRlciRncm91cF9iaXJ0aG9yZGVyLCB2YXIuZXF1YWwgPSBUKSkKY29oZW4uZChiaXJ0aG9yZGVyJHllYXJzX29mX2VkdWNhdGlvbiwgYXMuZmFjdG9yKGJpcnRob3JkZXIkZ3JvdXBfYmlydGhvcmRlciksIG5hLnJtID0gVCkKCgp0aWR5KHQudGVzdChiaXJ0aG9yZGVyJGJpZzVfZXh0IH4gYmlydGhvcmRlciRncm91cF9iaXJ0aG9yZGVyLCB2YXIuZXF1YWwgPSBUKSkKY29oZW4uZChiaXJ0aG9yZGVyJGJpZzVfZXh0LCBhcy5mYWN0b3IoYmlydGhvcmRlciRncm91cF9iaXJ0aG9yZGVyKSwgbmEucm0gPSBUKQoKCnRpZHkodC50ZXN0KGJpcnRob3JkZXIkYmlnNV9uZXUgfiBiaXJ0aG9yZGVyJGdyb3VwX2JpcnRob3JkZXIsIHZhci5lcXVhbCA9IFQpKQpjb2hlbi5kKGJpcnRob3JkZXIkYmlnNV9uZXUsIGFzLmZhY3RvcihiaXJ0aG9yZGVyJGdyb3VwX2JpcnRob3JkZXIpLCBuYS5ybSA9IFQpCgoKdGlkeSh0LnRlc3QoYmlydGhvcmRlciRiaWc1X2NvbiB+IGJpcnRob3JkZXIkZ3JvdXBfYmlydGhvcmRlciwgdmFyLmVxdWFsID0gVCkpCmNvaGVuLmQoYmlydGhvcmRlciRiaWc1X2NvbiwgYXMuZmFjdG9yKGJpcnRob3JkZXIkZ3JvdXBfYmlydGhvcmRlciksIG5hLnJtID0gVCkKCgp0aWR5KHQudGVzdChiaXJ0aG9yZGVyJGJpZzVfYWdyZWUgfiBiaXJ0aG9yZGVyJGdyb3VwX2JpcnRob3JkZXIsIHZhci5lcXVhbCA9IFQpKQpjb2hlbi5kKGJpcnRob3JkZXIkYmlnNV9hZ3JlZSwgYXMuZmFjdG9yKGJpcnRob3JkZXIkZ3JvdXBfYmlydGhvcmRlciksIG5hLnJtID0gVCkKCgp0aWR5KHQudGVzdChiaXJ0aG9yZGVyJGJpZzVfb3BlbiB+IGJpcnRob3JkZXIkZ3JvdXBfYmlydGhvcmRlciwgdmFyLmVxdWFsID0gVCkpCmNvaGVuLmQoYmlydGhvcmRlciRiaWc1X29wZW4sIGFzLmZhY3RvcihiaXJ0aG9yZGVyJGdyb3VwX2JpcnRob3JkZXIpLCBuYS5ybSA9IFQpCgoKdGlkeSh0LnRlc3QoYmlydGhvcmRlciRyaXNrQSB+IGJpcnRob3JkZXIkZ3JvdXBfYmlydGhvcmRlciwgdmFyLmVxdWFsID0gVCkpCmNvaGVuLmQoYmlydGhvcmRlciRyaXNrQSwgYXMuZmFjdG9yKGJpcnRob3JkZXIkZ3JvdXBfYmlydGhvcmRlciksIG5hLnJtID0gVCkKCnRpZHkodC50ZXN0KGJpcnRob3JkZXIkcmlza0IgfiBiaXJ0aG9yZGVyJGdyb3VwX2JpcnRob3JkZXIsIHZhci5lcXVhbCA9IFQpKQpjb2hlbi5kKGJpcnRob3JkZXIkcmlza0IsIGFzLmZhY3RvcihiaXJ0aG9yZGVyJGdyb3VwX2JpcnRob3JkZXIpLCBuYS5ybSA9IFQpCgojIyBFZHVjYXRpb25hbCBBdHRhaW5tZW50CmVkdWNhdGlvbmFsX2F0dGFpbm1lbnQgPSBiaXJ0aG9yZGVyICU+JQogIGdyb3VwX2J5KGdyb3VwX2JpcnRob3JkZXIpICU+JQogIHN1bW1hcmlzZSh5ZWFyc19vZl9lZHVjYXRpb25fbWVhbiA9IG1lYW4oeWVhcnNfb2ZfZWR1Y2F0aW9uLCBuYS5ybT1UKSwKICAgICAgICAgICAgeWVhcnNfb2ZfZWR1Y2F0aW9uX2NvbmZpZGVuY2VfbG93PSBtZWFuKHllYXJzX29mX2VkdWNhdGlvbiwgbmEucm0gPSBUKSAtIAogICAgICAgICAgICAgICAgKHF0KC45NzUsIG4oKS0xKSpzZCh5ZWFyc19vZl9lZHVjYXRpb24sIG5hLnJtID0gVCkvc3FydChuKCkpKSwKICAgICAgICAgICAgeWVhcnNfb2ZfZWR1Y2F0aW9uX2hpZ2ggPSBtZWFuKHllYXJzX29mX2VkdWNhdGlvbiwgbmEucm0gPSBUKSArCiAgICAgICAgICAgICAgICAocXQoLjk3NSwgbigpLTEpKnNkKHllYXJzX29mX2VkdWNhdGlvbiwgbmEucm0gPSBUKS9zcXJ0KG4oKSkpKQoKZWR1Y2F0aW9uYWxfYXR0YWlubWVudAoKdC50ZXN0KGJpcnRob3JkZXIkeWVhcnNfb2ZfZWR1Y2F0aW9uIH4gYmlydGhvcmRlciRncm91cF9iaXJ0aG9yZGVyLCB2YXIuZXF1YWwgPSBUKQoKZ2dwbG90KGRhdGE9YmlydGhvcmRlciwgYWVzKHg9eWVhcnNfb2ZfZWR1Y2F0aW9uLCBmaWxsPWdyb3VwX2JpcnRob3JkZXIpKSArCiAgZ2VvbV9oaXN0b2dyYW0oc3RhdD0iY291bnQiLCBiaW53aWR0aD0uNSwgcG9zaXRpb249ImRvZGdlIikKYGBgCgoKCiMjIERlc2NyaXB0aXZlcwpgYGB7ciBjb3JyZWxhdGlvbn0KeCA9IGJpcnRob3JkZXIgJT4lIGZpbHRlcihncm91cF9iaXJ0aG9yZGVyID09ICJ0ZXN0IiwgZ3JvdXBfb3V0Y29tZSA9PSAidGVzdCIpCm1lYW5fc2QgPSBiaXJ0aG9yZGVyICU+JQogIGdyb3VwX2J5KGdyb3VwX2JpcnRob3JkZXIpICU+JQogIHN1bW1hcmlzZShhZ2VfbWVhbiA9IG1lYW4oYWdlLCBuYS5ybT1UKSwKICAgICAgICAgICAgYWdlX3NkID0gc2QoYWdlLCBuYS5ybT1UKSwKICAgICAgICAgICAgZ19mYWN0b3JfbWVhbiA9IG1lYW4oZ19mYWN0b3JfMjAxNV9vbGQsIG5hLnJtPVQpLAogICAgICAgICAgICBnX2ZhY3Rvcl9zZCA9IHNkKGdfZmFjdG9yXzIwMTVfb2xkLCBuYS5ybT1UKSwKICAgICAgICAgICAgYmlnNV9leHRfbWVhbiA9IG1lYW4oYmlnNV9leHQsIG5hLnJtPVQpLAogICAgICAgICAgICBiaWc1X2V4dF9zZCA9IHNkKGJpZzVfZXh0LCBuYS5ybT1UKSwKICAgICAgICAgICAgYmlnNV9uZXVfbWVhbiA9IG1lYW4oYmlnNV9uZXUsIG5hLnJtPVQpLAogICAgICAgICAgICBiaWc1X25ldV9zZCA9IHNkKGJpZzVfbmV1LCBuYS5ybT1UKSwKICAgICAgICAgICAgYmlnNV9jb25fbWVhbiA9IG1lYW4oYmlnNV9jb24sIG5hLnJtPVQpLAogICAgICAgICAgICBiaWc1X2Nvbl9zZCA9IHNkKGJpZzVfY29uLCBuYS5ybT1UKSwKICAgICAgICAgICAgYmlnNV9hZ3JlZV9tZWFuID0gbWVhbihiaWc1X2FncmVlLCBuYS5ybT1UKSwKICAgICAgICAgICAgYmlnNV9hZ3JlZV9zZCA9IHNkKGJpZzVfYWdyZWUsIG5hLnJtPVQpLAogICAgICAgICAgICBiaWc1X29wZW5fbWVhbiA9IG1lYW4oYmlnNV9vcGVuLCBuYS5ybT1UKSwKICAgICAgICAgICAgYmlnNV9vcGVuX3NkID0gc2QoYmlnNV9vcGVuLCBuYS5ybT1UKSwKICAgICAgICAgICAgcmlza0FfbWVhbiA9IG1lYW4ocmlza0EsIG5hLnJtPVQpLAogICAgICAgICAgICByaXNrQV9zZCA9IHNkKHJpc2tBLCBuYS5ybT1UKSwKICAgICAgICAgICAgcmlza0JfbWVhbiA9IG1lYW4ocmlza0IsIG5hLnJtPVQpLAogICAgICAgICAgICByaXNrQl9zZCA9IHNkKHJpc2tCLCBuYS5ybT1UKSwKICAgICAgICAgICAgeWVhcnNfb2ZfZWR1Y2F0aW9uX21lYW4gPSBtZWFuKHllYXJzX29mX2VkdWNhdGlvbiwgbmEucm09VCksCiAgICAgICAgICAgIHllYXJzX29mX2VkdWNhdGlvbl9zZCA9IHNkKHllYXJzX29mX2VkdWNhdGlvbiwgbmEucm09VCkpCiAgICAgIAptZWFuX3NkCmBgYAoKIyMgQ29ycmVsYXRpb25zCmBgYHtyfQpjb3IgPSByb3VuZChjb3IoYmlydGhvcmRlciAlPiUgZmlsdGVyKGdyb3VwX2JpcnRob3JkZXIgPT0gInRlc3QiLCBncm91cF9vdXRjb21lID09ICJ0ZXN0IikgJT4lIHVuZ3JvdXAoKSAlPiUgc2VsZWN0KGFnZSwgbWFsZSwgZ19mYWN0b3JfMjAxNV9vbGQsIHllYXJzX29mX2VkdWNhdGlvbiwgYmlnNV9leHQsIGJpZzVfbmV1LCBiaWc1X2NvbiwgYmlnNV9hZ3JlZSwgYmlnNV9vcGVuLCByaXNrQSwgcmlza0IpLCB1c2UgPSAicGFpcndpc2UuY29tcGxldGUub2JzIiksIDIpIAoKY29yCmBgYAoKIyMgUmVsaWFiaWxpdHkgey50YWJzZXR9CgojIyMgSW50ZWxsaWdlbmNlCmBgYHtyfQphbHBoYShiaXJ0aG9yZGVyICU+JSBmaWx0ZXIoZ3JvdXBfYmlydGhvcmRlciA9PSAidGVzdCIsIGdyb3VwX291dGNvbWUgPT0gInRlc3QiKSAlPiUgdW5ncm91cCgpICU+JSBzZWxlY3QocmF2ZW5fMjAxNV9vbGQsIG1hdGhfMjAxNV9vbGQsIGNvdW50X2JhY2t3YXJkcywgd29yZHNfZGVsYXllZCwgYWRhcHRpdmVfbnVtYmVyaW5nKSkKYGBgCgojIyMgUGVyc29uYWxpdHkKYGBge3J9CiMjRXh0cmF2ZXJzaW9uCmFscGhhKGJpcnRob3JkZXIgJT4lIGZpbHRlcihncm91cF9iaXJ0aG9yZGVyID09ICJ0ZXN0IiwgZ3JvdXBfb3V0Y29tZSA9PSAidGVzdCIpICU+JSB1bmdyb3VwKCkgJT4lCiAgICAgICAgc2VsZWN0KGUxLCBlMnJfcmV2ZXJzZWQsIGUzKSkKCiMjIE5ldXJvdGljaXNtCmFscGhhKGJpcnRob3JkZXIgJT4lIGZpbHRlcihncm91cF9iaXJ0aG9yZGVyID09ICJ0ZXN0IiwgZ3JvdXBfb3V0Y29tZSA9PSAidGVzdCIpICU+JSB1bmdyb3VwKCkKICAgICAgJT4lIHNlbGVjdChuMXJfcmV2ZXJzZWQsIG4yLCBuMykpCgojI2NvbnNjaWVudGlvdXNuZXNzCmFscGhhKGJpcnRob3JkZXIgJT4lIGZpbHRlcihncm91cF9iaXJ0aG9yZGVyID09ICJ0ZXN0IiwgZ3JvdXBfb3V0Y29tZSA9PSAidGVzdCIpICU+JSB1bmdyb3VwKCkgJT4lIHNlbGVjdChjMSwgYzJyX3JldmVyc2VkLCBjMykpCgojI0FncmVlYWJsZW5lc3MKYWxwaGEoYmlydGhvcmRlciAlPiUgZmlsdGVyKGdyb3VwX2JpcnRob3JkZXIgPT0gInRlc3QiLCBncm91cF9vdXRjb21lID09ICJ0ZXN0IikgJT4lIHVuZ3JvdXAoKSAlPiUgc2VsZWN0KGExLCBhMiwgYTNyX3JldmVyc2VkKSkKCiMjT3Blbm5lc3MKYWxwaGEoYmlydGhvcmRlciAlPiUgZmlsdGVyKGdyb3VwX2JpcnRob3JkZXIgPT0gInRlc3QiLCBncm91cF9vdXRjb21lID09ICJ0ZXN0IikgJT4lIHVuZ3JvdXAoKSAlPiUgc2VsZWN0KG8xLCBvMiwgbzMpKQpgYGAKCgojIyBQbG90cyBhZ2UgYW5kIGdlbmRlciB7LnRhYnNldH0KCiMjIyBQbG90cyBHZW5kZXIKYGBge3J9CmJpcnRob3JkZXIgPSBiaXJ0aG9yZGVyICU+JSBmaWx0ZXIoYWdlPD0xMDApCgpiaXJ0aG9yZGVyID0gYmlydGhvcmRlciAlPiUKICBtdXRhdGUob3V0Y29tZSA9IGFnZSkKcGxvdF9nZW5kZXIoYmlydGhvcmRlcikKCmJpcnRob3JkZXIgPSBiaXJ0aG9yZGVyICU+JQogIG11dGF0ZShvdXRjb21lID0gYmlydGhvcmRlcl9nZW5lcykKcGxvdF9nZW5kZXIoYmlydGhvcmRlcikKCmJpcnRob3JkZXIgPSBiaXJ0aG9yZGVyICU+JQogIG11dGF0ZShvdXRjb21lID0gc2libGluZ19jb3VudF9nZW5lcykKcGxvdF9nZW5kZXIoYmlydGhvcmRlcikKCgpiaXJ0aG9yZGVyID0gYmlydGhvcmRlciAlPiUKICBtdXRhdGUob3V0Y29tZSA9IGdfZmFjdG9yXzIwMTVfb2xkKQpwbG90X2dlbmRlcihiaXJ0aG9yZGVyKQoKYmlydGhvcmRlciA9IGJpcnRob3JkZXIgJT4lCiAgbXV0YXRlKG91dGNvbWUgPSBnX2ZhY3Rvcl8yMDE1X3lvdW5nKQpwbG90X2dlbmRlcihiaXJ0aG9yZGVyKQoKYmlydGhvcmRlciA9IGJpcnRob3JkZXIgJT4lCiAgbXV0YXRlKG91dGNvbWUgPSBnX2ZhY3Rvcl8yMDA3X29sZCkKcGxvdF9nZW5kZXIoYmlydGhvcmRlcikKCmJpcnRob3JkZXIgPSBiaXJ0aG9yZGVyICU+JQogIG11dGF0ZShvdXRjb21lID0gZ19mYWN0b3JfMjAwN195b3VuZykKcGxvdF9nZW5kZXIoYmlydGhvcmRlcikKCmJpcnRob3JkZXIgPSBiaXJ0aG9yZGVyICU+JQogIG11dGF0ZShvdXRjb21lID0gYmlnNV9leHQpCnBsb3RfZ2VuZGVyKGJpcnRob3JkZXIpCgpiaXJ0aG9yZGVyID0gYmlydGhvcmRlciAlPiUKICBtdXRhdGUob3V0Y29tZSA9IGJpZzVfY29uKQpwbG90X2dlbmRlcihiaXJ0aG9yZGVyKQoKYmlydGhvcmRlciA9IGJpcnRob3JkZXIgJT4lCiAgbXV0YXRlKG91dGNvbWUgPSBiaWc1X29wZW4pCnBsb3RfZ2VuZGVyKGJpcnRob3JkZXIpCgpiaXJ0aG9yZGVyID0gYmlydGhvcmRlciAlPiUKICBtdXRhdGUob3V0Y29tZSA9IGJpZzVfbmV1KQpwbG90X2dlbmRlcihiaXJ0aG9yZGVyKQoKYmlydGhvcmRlciA9IGJpcnRob3JkZXIgJT4lCiAgbXV0YXRlKG91dGNvbWUgPSBiaWc1X2FncmVlKQpwbG90X2dlbmRlcihiaXJ0aG9yZGVyKQoKYmlydGhvcmRlciA9IGJpcnRob3JkZXIgJT4lCiAgbXV0YXRlKG91dGNvbWUgPSByaXNrQSkKcGxvdF9nZW5kZXIoYmlydGhvcmRlcikKCmJpcnRob3JkZXIgPSBiaXJ0aG9yZGVyICU+JQogIG11dGF0ZShvdXRjb21lID0gcmlza0IpCnBsb3RfZ2VuZGVyKGJpcnRob3JkZXIpCgpiaXJ0aG9yZGVyID0gYmlydGhvcmRlciAlPiUKICBtdXRhdGUob3V0Y29tZSA9IHllYXJzX29mX2VkdWNhdGlvbikKcGxvdF9nZW5kZXIoYmlydGhvcmRlcikKCnBsb3RfZ2VuZGVyKGJpcnRob3JkZXIgJT4lIGZpbHRlcighaXMubmEoYXR0ZW5kZWRfc2Nob29sKSkgJT4lIG11dGF0ZShvdXRjb21lID0gYXMubnVtZXJpYyhhdHRlbmRlZF9zY2hvb2wpKSkKCmJpcnRob3JkZXIgPSBiaXJ0aG9yZGVyICU+JQogIG11dGF0ZShvdXRjb21lID0gRWxlbWVudGFyeV9taXNzZWQpCnBsb3RfZ2VuZGVyKGJpcnRob3JkZXIpCgpiaXJ0aG9yZGVyID0gYmlydGhvcmRlciAlPiUKICBtdXRhdGUob3V0Y29tZSA9IEVsZW1lbnRhcnlfd29ya2VkKQpwbG90X2dlbmRlcihiaXJ0aG9yZGVyKQoKYmlydGhvcmRlciA9IGJpcnRob3JkZXIgJT4lCiAgbXV0YXRlKG91dGNvbWUgPSB3YWdlX2xhc3RfbW9udGhfbG9nKQpwbG90X2dlbmRlcihiaXJ0aG9yZGVyKQoKYmlydGhvcmRlciA9IGJpcnRob3JkZXIgJT4lCiAgbXV0YXRlKG91dGNvbWUgPSB3YWdlX2xhc3RfeWVhcl9sb2cpCnBsb3RfZ2VuZGVyKGJpcnRob3JkZXIpCgpiaXJ0aG9yZGVyID0gYmlydGhvcmRlciAlPiUKICBtdXRhdGUob3V0Y29tZSA9IFNlbGZfZW1wbG95ZWQpCnBsb3RfZ2VuZGVyKGJpcnRob3JkZXIpCgpiaXJ0aG9yZGVyID0gYmlydGhvcmRlciAlPiUKICBtdXRhdGUob3V0Y29tZSA9IGV2ZXJfc21va2VkKQpwbG90X2dlbmRlcihiaXJ0aG9yZGVyKQoKYGBgCgojIyMgUGxvdHMgQWdlCmBgYHtyfQpwbG90X2FnZShiaXJ0aG9yZGVyICU+JSBmaWx0ZXIoYWdlPD0gMTAwKSAlPiUgbXV0YXRlKG91dGNvbWUgPSBiaXJ0aG9yZGVyX2dlbmVzKSkKCgpiaXJ0aG9yZGVyID0gYmlydGhvcmRlciAlPiUKICBtdXRhdGUob3V0Y29tZSA9IHNpYmxpbmdfY291bnRfZ2VuZXMpCnBsb3RfYWdlKGJpcnRob3JkZXIpCgoKYmlydGhvcmRlciA9IGJpcnRob3JkZXIgJT4lCiAgbXV0YXRlKG91dGNvbWUgPSBnX2ZhY3Rvcl8yMDE1X29sZCkKcGxvdF9hZ2UoYmlydGhvcmRlcikKCmJpcnRob3JkZXIgPSBiaXJ0aG9yZGVyICU+JQogIG11dGF0ZShvdXRjb21lID0gZ19mYWN0b3JfMjAxNV95b3VuZykKcGxvdF9hZ2UoYmlydGhvcmRlcikKCmJpcnRob3JkZXIgPSBiaXJ0aG9yZGVyICU+JQogIG11dGF0ZShvdXRjb21lID0gZ19mYWN0b3JfMjAwN19vbGQpCnBsb3RfYWdlKGJpcnRob3JkZXIpCgpiaXJ0aG9yZGVyID0gYmlydGhvcmRlciAlPiUKICBtdXRhdGUob3V0Y29tZSA9IGdfZmFjdG9yXzIwMDdfeW91bmcpCnBsb3RfYWdlKGJpcnRob3JkZXIpCgpiaXJ0aG9yZGVyID0gYmlydGhvcmRlciAlPiUKICBtdXRhdGUob3V0Y29tZSA9IGJpZzVfZXh0KQpwbG90X2FnZShiaXJ0aG9yZGVyKQoKYmlydGhvcmRlciA9IGJpcnRob3JkZXIgJT4lCiAgbXV0YXRlKG91dGNvbWUgPSBiaWc1X2NvbikKcGxvdF9hZ2UoYmlydGhvcmRlcikKCmJpcnRob3JkZXIgPSBiaXJ0aG9yZGVyICU+JQogIG11dGF0ZShvdXRjb21lID0gYmlnNV9vcGVuKQpwbG90X2FnZShiaXJ0aG9yZGVyKQoKYmlydGhvcmRlciA9IGJpcnRob3JkZXIgJT4lCiAgbXV0YXRlKG91dGNvbWUgPSBiaWc1X25ldSkKcGxvdF9hZ2UoYmlydGhvcmRlcikKCmJpcnRob3JkZXIgPSBiaXJ0aG9yZGVyICU+JQogIG11dGF0ZShvdXRjb21lID0gYmlnNV9hZ3JlZSkKcGxvdF9hZ2UoYmlydGhvcmRlcikKCmJpcnRob3JkZXIgPSBiaXJ0aG9yZGVyICU+JQogIG11dGF0ZShvdXRjb21lID0gcmlza0EpCnBsb3RfYWdlKGJpcnRob3JkZXIpCgpiaXJ0aG9yZGVyID0gYmlydGhvcmRlciAlPiUKICBtdXRhdGUob3V0Y29tZSA9IHJpc2tCKQpwbG90X2FnZShiaXJ0aG9yZGVyKQoKYmlydGhvcmRlciA9IGJpcnRob3JkZXIgJT4lCiAgbXV0YXRlKG91dGNvbWUgPSB5ZWFyc19vZl9lZHVjYXRpb24pCnBsb3RfYWdlKGJpcnRob3JkZXIpCgpwbG90X2FnZShiaXJ0aG9yZGVyICU+JSBmaWx0ZXIoIWlzLm5hKGF0dGVuZGVkX3NjaG9vbCkpICU+JSBtdXRhdGUob3V0Y29tZSA9IGFzLm51bWVyaWMoYXR0ZW5kZWRfc2Nob29sKSkpCgpiaXJ0aG9yZGVyID0gYmlydGhvcmRlciAlPiUKICBtdXRhdGUob3V0Y29tZSA9IEVsZW1lbnRhcnlfbWlzc2VkKQpwbG90X2FnZShiaXJ0aG9yZGVyKQoKYmlydGhvcmRlciA9IGJpcnRob3JkZXIgJT4lCiAgbXV0YXRlKG91dGNvbWUgPSBFbGVtZW50YXJ5X3dvcmtlZCkKcGxvdF9hZ2UoYmlydGhvcmRlcikKCmJpcnRob3JkZXIgPSBiaXJ0aG9yZGVyICU+JQogIG11dGF0ZShvdXRjb21lID0gd2FnZV9sYXN0X21vbnRoX2xvZykKcGxvdF9hZ2UoYmlydGhvcmRlcikKCmJpcnRob3JkZXIgPSBiaXJ0aG9yZGVyICU+JQogIG11dGF0ZShvdXRjb21lID0gd2FnZV9sYXN0X3llYXJfbG9nKQpwbG90X2FnZShiaXJ0aG9yZGVyKQoKYmlydGhvcmRlciA9IGJpcnRob3JkZXIgJT4lCiAgbXV0YXRlKG91dGNvbWUgPSBTZWxmX2VtcGxveWVkKQpwbG90X2FnZShiaXJ0aG9yZGVyKQoKYmlydGhvcmRlciA9IGJpcnRob3JkZXIgJT4lCiAgbXV0YXRlKG91dGNvbWUgPSBldmVyX3Ntb2tlZCkKcGxvdF9hZ2UoYmlydGhvcmRlcikKCmBgYAoK