Codebook
source("0_helpers.R")
birthorder = readRDS("data/alldata_birthorder.rds")
knitr::opts_chunk$set(error = TRUE, warning = F, message = F)Data
# For analyses we want to clean the dataset and get rid of all uninteresting data
birthorder = birthorder %>%
mutate(money_spent_smoking_log = if_else(is.na(money_spent_smoking_log) & ever_smoked == 0, 0, money_spent_smoking_log),
amount = if_else(is.na(amount) & ever_smoked == 0, 0, amount),
amount_still_smokers = if_else(is.na(amount_still_smokers) & still_smoking == 0, 0, amount_still_smokers),
birthyear = lubridate::year(birthdate))
### Variables
birthorder = birthorder %>%
mutate(
attended_school = as.integer(attended_school),
attended_school = ifelse(attended_school == 1, 0,
ifelse(attended_school == 2, 1, NA)))
### Birthorder and Sibling Count
birthorder = birthorder %>%
mutate(
# birthorder as factors with levels of 1, 2, 3, 4, 5, >5
birthorder_naive_factor = as.character(birthorder_naive),
birthorder_naive_factor = ifelse(birthorder_naive > 5, ">5",
birthorder_naive_factor),
birthorder_naive_factor = factor(birthorder_naive_factor,
levels = c("1","2","3","4","5",">5")),
sibling_count_naive_factor = as.character(sibling_count_naive),
sibling_count_naive_factor = ifelse(sibling_count_naive > 5, ">5",
sibling_count_naive_factor),
sibling_count_naive_factor = factor(sibling_count_naive_factor,
levels = c("2","3","4","5",">5")),
birthorder_uterus_alive_factor = as.character(birthorder_uterus_alive),
birthorder_uterus_alive_factor = ifelse(birthorder_uterus_alive > 5, ">5",
birthorder_uterus_alive_factor),
birthorder_uterus_alive_factor = factor(birthorder_uterus_alive_factor,
levels = c("1","2","3","4","5",">5")),
sibling_count_uterus_alive_factor = as.character(sibling_count_uterus_alive),
sibling_count_uterus_alive_factor = ifelse(sibling_count_uterus_alive > 5, ">5",
sibling_count_uterus_alive_factor),
sibling_count_uterus_alive_factor = factor(sibling_count_uterus_alive_factor,
levels = c("2","3","4","5",">5")),
birthorder_uterus_preg_factor = as.character(birthorder_uterus_preg),
birthorder_uterus_preg_factor = ifelse(birthorder_uterus_preg > 5, ">5",
birthorder_uterus_preg_factor),
birthorder_uterus_preg_factor = factor(birthorder_uterus_preg_factor,
levels = c("1","2","3","4","5",">5")),
sibling_count_uterus_preg_factor = as.character(sibling_count_uterus_preg),
sibling_count_uterus_preg_factor = ifelse(sibling_count_uterus_preg > 5, ">5",
sibling_count_uterus_preg_factor),
sibling_count_uterus_preg_factor = factor(sibling_count_uterus_preg_factor,
levels = c("2","3","4","5",">5")),
birthorder_genes_factor = as.character(birthorder_genes),
birthorder_genes_factor = ifelse(birthorder_genes >5 , ">5", birthorder_genes_factor),
birthorder_genes_factor = factor(birthorder_genes_factor,
levels = c("1","2","3","4","5",">5")),
sibling_count_genes_factor = as.character(sibling_count_genes),
sibling_count_genes_factor = ifelse(sibling_count_genes >5 , ">5",
sibling_count_genes_factor),
sibling_count_genes_factor = factor(sibling_count_genes_factor,
levels = c("2","3","4","5",">5")),
# interaction birthorder * siblingcout for each birthorder
count_birthorder_naive =
factor(str_replace(as.character(interaction(birthorder_naive_factor, sibling_count_naive_factor)),
"\\.", "/"),
levels = c("1/2","2/2", "1/3", "2/3",
"3/3", "1/4", "2/4", "3/4", "4/4",
"1/5", "2/5", "3/5", "4/5", "5/5",
"1/>5", "2/>5", "3/>5", "4/>5",
"5/>5", ">5/>5")),
count_birthorder_uterus_alive =
factor(str_replace(as.character(interaction(birthorder_uterus_alive_factor, sibling_count_uterus_alive_factor)),
"\\.", "/"),
levels = c("1/2","2/2", "1/3", "2/3",
"3/3", "1/4", "2/4", "3/4", "4/4",
"1/5", "2/5", "3/5", "4/5", "5/5",
"1/>5", "2/>5", "3/>5", "4/>5",
"5/>5", ">5/>5")),
count_birthorder_uterus_preg =
factor(str_replace(as.character(interaction(birthorder_uterus_preg_factor, sibling_count_uterus_preg_factor)),
"\\.", "/"),
levels = c("1/2","2/2", "1/3", "2/3",
"3/3", "1/4", "2/4", "3/4", "4/4",
"1/5", "2/5", "3/5", "4/5", "5/5",
"1/>5", "2/>5", "3/>5", "4/>5",
"5/>5", ">5/>5")),
count_birthorder_genes =
factor(str_replace(as.character(interaction(birthorder_genes_factor, sibling_count_genes_factor)), "\\.", "/"),
levels = c("1/2","2/2", "1/3", "2/3",
"3/3", "1/4", "2/4", "3/4", "4/4",
"1/5", "2/5", "3/5", "4/5", "5/5",
"1/>5", "2/>5", "3/>5", "4/>5",
"5/>5", ">5/>5")))
birthorder <- birthorder %>%
mutate(sibling_count = sibling_count_naive_factor,
birth_order_nonlinear = birthorder_naive_factor,
birth_order = birthorder_naive,
count_birth_order = count_birthorder_naive)birthorder$mother_pidlink <- as.character(birthorder$mother_pidlink)
birthorder$pidlink <- as.character(birthorder$pidlink)
birthorder$father_pidlink <- as.character(birthorder$father_pidlink)
birthorder$marriage_id <- as.character(birthorder$marriage_id)
library(codebook)
var_label(birthorder$e1) <- "Is talkative"
var_label(birthorder$c1) <- "Does a thorough job"
var_label(birthorder$o1) <- "Is original, comes up with new ideas."
var_label(birthorder$e2r_reversed) <- "Is reserved."
var_label(birthorder$n1r_reversed) <- "Is relaxed, handles stress well."
var_label(birthorder$a1) <- "Has a forgiving nature."
var_label(birthorder$n2) <- "Worries a lot."
var_label(birthorder$o2) <- "Has an active imagination."
var_label(birthorder$c2r_reversed) <- "Tends to be lazy."
var_label(birthorder$o3) <- "Values artistic, aesthetic experiences."
var_label(birthorder$a2) <- "Is considerate and kind to almost everyone."
var_label(birthorder$c3) <- "Does things efficiently."
var_label(birthorder$e3) <- "Outgoing, sociable."
var_label(birthorder$a3r_reversed) <- "Is sometimes rude to others."
var_label(birthorder$n3) <- "Gets nervous easily."
add_likert_labels <- function(x) {
val_labels(x) <- c("Disagree strongly" = 1,
"Disagree a little" = 2,
"Neither agree nor disagree" = 3,
"Agree a little" = 4,
"Agree strongly" = 5)
x
}
birthorder <- birthorder %>% mutate_at(vars(e1, c1, o1, e2r, n1r, a1, n2, o2, c2r, o3, a2, c3, e3, a3r, n3), add_likert_labels)
##Extraversion
birthorder$e2r_reversed = codebook::reverse_labelled_values(birthorder$e2r)
extraversion = birthorder %>% select(e1, e2r_reversed, e3)
birthorder$big5_ext = aggregate_and_document_scale(extraversion)
##conscientiousness
birthorder$c2r_reversed = codebook::reverse_labelled_values(birthorder$c2r)
conscientiousness = birthorder %>% select(c1, c2r_reversed, c3)
birthorder$big5_con = aggregate_and_document_scale(conscientiousness)
##Openness
openness = birthorder %>% select(o1, o2, o3)
birthorder$big5_open = aggregate_and_document_scale(openness)
## Neuroticism
birthorder$n1r_reversed = codebook::reverse_labelled_values(birthorder$n1r)
neuroticism = birthorder %>% select(n1r_reversed, n2, n3)
birthorder$big5_neu = aggregate_and_document_scale(neuroticism)
##Agreeableness
birthorder$a3r_reversed = codebook::reverse_labelled_values(birthorder$a3r)
agreeableness= birthorder %>% select(a1, a2, a3r_reversed)
birthorder$big5_agree = aggregate_and_document_scale(agreeableness)
cb_table <- codebook_table(birthorder)
rio::export(cb_table, "2_codebook.xlsx")
metadata(birthorder)$name <- "Indonesian Family Life Study, merged subset"
metadata(birthorder)$description <- "Data from the IFLS, merged across waves, most outcomes taken from wave 5. Includes birth order, family structure, Big 5 Personality, intelligence tests, and risk lotteries"
metadata(birthorder)$identifier <- "https://www.rand.org/well-being/social-and-behavioral-policy/data/FLS/IFLS.html"
metadata(birthorder)$creator <- "RAND corporation"
metadata(birthorder)$citation <- "Strauss, J., Witoelar, F., & Sikoki, B. (2016). The Fifth Wave of the Indonesia Family Life Survey: Overview and Field Report. WR-1143/1-NIA/NICHD"
metadata(birthorder)$url <- "https://www.rand.org/well-being/social-and-behavioral-policy/data/FLS/IFLS.html"
metadata(birthorder)$datePublished <- "2016"
metadata(birthorder)$temporalCoverage <- "2014/2015"
metadata(birthorder)$spatialCoverage <- "13 Indonesian provinces. The sample is representative of about 83% of the Indonesian population and contains over 30,000 individuals living in 13 of the 27 provinces in the country. See URL for more." codebook(birthorder, survey_repetition = "single",
detailed_variables = FALSE, detailed_scales = TRUE, missingness_report = FALSE,
metadata_table = TRUE, metadata_json = TRUE, indent = "#")knitr::asis_output(data_info)Metadata
Description
if (exists("name", meta)) {
glue::glue(
"__Dataset name__: {name}",
.envir = meta)
}Dataset name: Indonesian Family Life Study, merged subset
cat(description)Data from the IFLS, merged across waves, most outcomes taken from wave 5. Includes birth order, family structure, Big 5 Personality, intelligence tests, and risk lotteries
Metadata for search engines
- Temporal Coverage: 2014/2015
- Spatial Coverage: 13 Indonesian provinces. The sample is representative of about 83% of the Indonesian population and contains over 30,000 individuals living in 13 of the 27 provinces in the country. See URL for more.
- Citation: Strauss, J., Witoelar, F., & Sikoki, B. (2016). The Fifth Wave of the Indonesia Family Life Survey: Overview and Field Report. WR-1143/1-NIA/NICHD
- URL: https://www.rand.org/well-being/social-and-behavioral-policy/data/FLS/IFLS.html
- Identifier: https://www.rand.org/well-being/social-and-behavioral-policy/data/FLS/IFLS.html
Date published: 2016
Creator:RAND corporation
meta <- meta[setdiff(names(meta),
c("creator", "datePublished", "identifier",
"url", "citation", "spatialCoverage",
"temporalCoverage", "description", "name"))]
pander::pander(meta)- keywords: wave, mother_pidlink, chron_order_birth, lifebirths, multiple_birth, alive, birthdate, any_multiple_birth, marriage_id, birthorder_uterus_preg, sibling_count_uterus_preg, birthorder_uterus_alive, sibling_count_uterus_alive, birthorder_genes, sibling_count_genes, pidlink, father_pidlink, age, death_yr, death_month, sc05, province, sc01_14_14, sibling_count_naive_ind, any_multiple_birthdate, birthorder_naive, sibling_count_naive, age_2015_old, age_2015_young, raven_2015_young, math_2015_young, raven_2015_old, math_2015_old, words_immediate, words_delayed, words_remembered_avg, adaptive_numbering, age_2007_young, age_2007_old, raven_2007_old, raven_2007_young, math_2007_young, math_2007_old, count_backwards, g_factor_2015_old, g_factor_2015_young, g_factor_2007_old, g_factor_2007_young, e1, c1, o1, e2r, n1r, a1, n2, o2, c2r, o3, a2, c3, e3, a3r, n3, e2r_reversed, big5_ext, c2r_reversed, big5_con, big5_open, n1r_reversed, big5_neu, a3r_reversed, big5_agree, random_si, si01, si02, si03, si04, si05, si11, si12, si13, si14, si15, riskA, riskB, attended_school, highest_education, currently_attending_school, hours_in_class, years_of_education, Type_of_test_elementary, Indonesia_score_elementary, English_score_elementary, Math_score_elemenatry, Total_score_elemenatry, Type_of_test_Junior_High, Indonesia_score_Junior_High, English_score_Junior_High, Math_score_Junior_High, Total_score_Junior_High, Type_of_test_Senior_High, Indonesia_score_Senior_High, English_score_Senior_High, Math_score_Senior_High, Total_score_Senior_High, Total_score_highest, Total_score_highest_type, Math_score_highest, Math_score_highest_type, Elementary_worked, Junior_high_worked, Senior_high_worked, University_worked, total_worked, Elementary_missed, Junior_high_missed, Senior_high_missed, University_missed, total_missed, Category, Sector, Self_employed, ever_smoked, still_smoking, amount, age_first_smoke, amount_still_smokers, male, wage_last_month_log, wage_last_year_log, money_spent_smoking_log, birthyear, birthorder_naive_factor, sibling_count_naive_factor, birthorder_uterus_alive_factor, sibling_count_uterus_alive_factor, birthorder_uterus_preg_factor, sibling_count_uterus_preg_factor, birthorder_genes_factor, sibling_count_genes_factor, count_birthorder_naive, count_birthorder_uterus_alive, count_birthorder_uterus_preg, count_birthorder_genes, sibling_count, birth_order_nonlinear, birth_order and count_birth_order
knitr::asis_output(survey_overview)Variables
if (detailed_variables || detailed_scales) {
knitr::asis_output(paste0(scales_items, sep = "\n\n\n", collapse = "\n\n\n"))
}Scale: big5_ext
Overview
Reliability: ωordinal [95% CI] = 0.59 [0.52;0.65].
Missing: 69795.
old_height <- knitr::opts_chunk$get("fig.height")
new_height <- length(scale_info$scale_item_names)
new_height <- ifelse(new_height > 20, 20, new_height)
new_height <- ifelse(new_height < 1, 1, new_height)
new_height <- ifelse(is.na(new_height) | is.nan(new_height),
old_height, new_height)
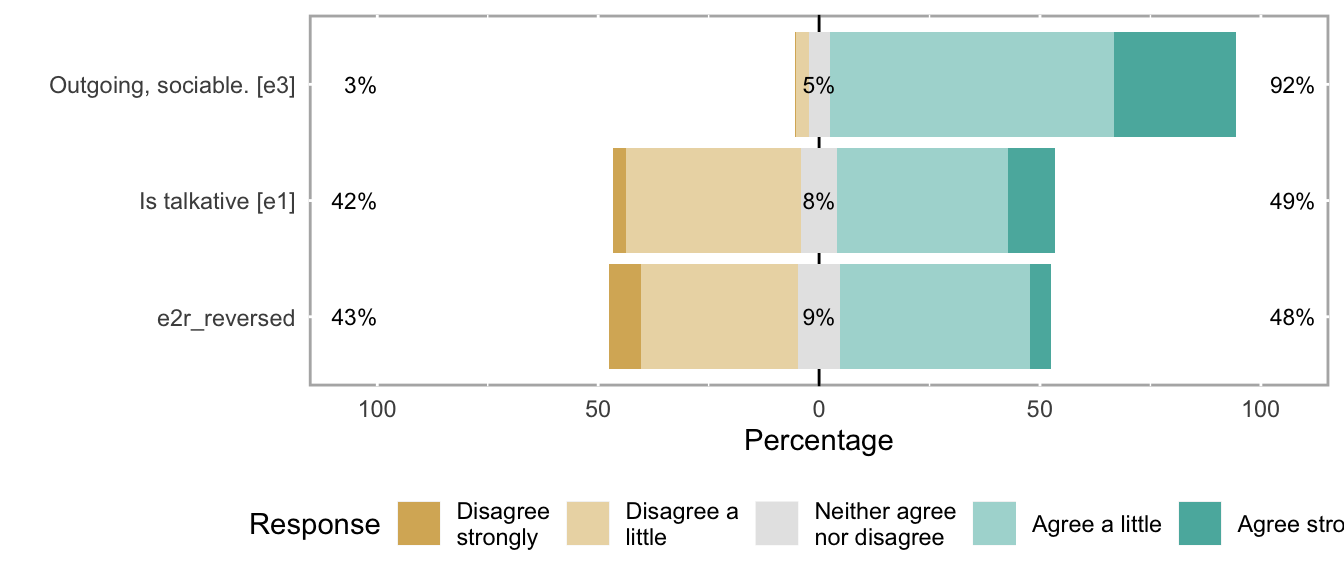
knitr::opts_chunk$set(fig.height = new_height)if (dplyr::n_distinct(na.omit(unlist(items))) < 12) {
likert_plot <- likert_from_items(items)
if (!is.null(likert_plot)) {
graphics::plot(likert_plot)
}
}
knitr::opts_chunk$set(fig.height = old_height)wrap_at <- knitr::opts_chunk$get("fig.width") * 10dist_plot <- plot_labelled(scale, scale_name, wrap_at)
choices <- attributes(items[[1]])$item$choices
breaks <- as.numeric(names(choices))
if (length(breaks)) {
suppressMessages( # ignore message about overwriting x axis
dist_plot <- dist_plot +
ggplot2::scale_x_continuous("values",
breaks = breaks,
labels = stringr::str_wrap(unlist(choices), ceiling(wrap_at * 0.21))) +
ggplot2::expand_limits(x = range(breaks)))
}
dist_plot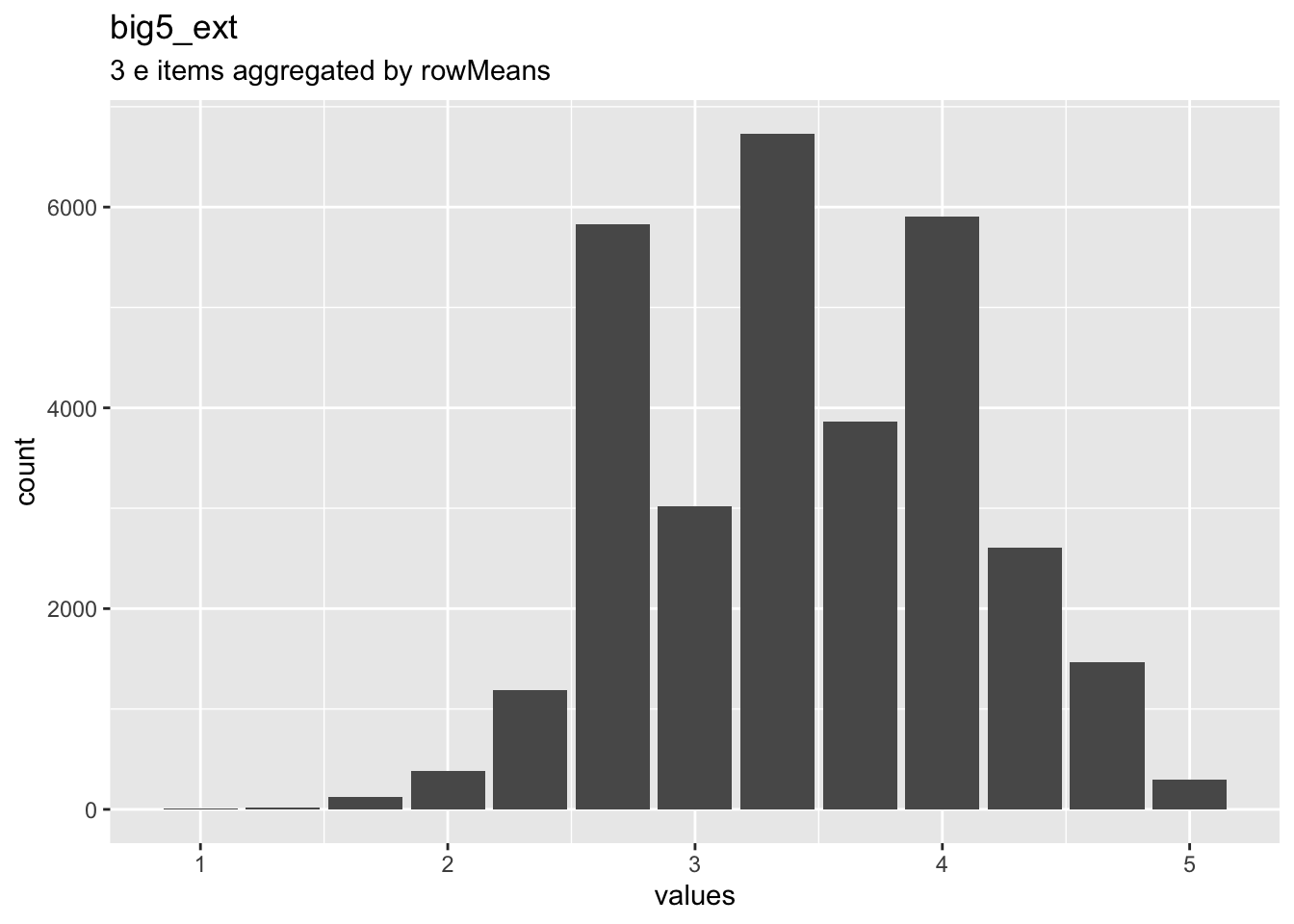
Reliability details
for (i in seq_along(reliabilities)) {
rel <- reliabilities[[i]]
cat(knitr::knit_print(rel, indent = paste0(indent, "####")))
}Reliability Indices
coefs <- x$scaleReliability$output$dat %>%
tidyr::gather(index, estimate) %>%
dplyr::filter(index != "n.items", index != "n.observations") %>%
dplyr::mutate(index = stringr::str_to_title(
stringr::str_replace_all(index,
stringr::fixed("."), " ")))
cis <- coefs %>%
dplyr::filter(stringr::str_detect(index, " Ci ")) %>%
tidyr::separate(index, c("index", "hilo"), sep = " Ci ") %>%
tidyr::spread(hilo, estimate)
if (nrow(cis)) {
cis <- cis %>% dplyr::rename(
`Lower 95% CI` = .data$Lo, `Upper 95% CI` = .data$Hi
)
}
coefs_with_cis <- coefs %>%
dplyr::filter(!stringr::str_detect(index, " Ci ")) %>%
dplyr::left_join(cis, by = "index") %>%
dplyr::mutate(index = dplyr::if_else(index == "Glb", "Greatest Lower Bound", .data$index)) %>%
dplyr::arrange(!stringr::str_detect(index, "Omega")) %>%
dplyr::select(Index = .data$index, Estimate = .data$estimate)
pander::pander(coefs_with_cis)| Index | Estimate |
|---|---|
| Omega | 0.5702 |
| Omega Psych Tot | 0.2915 |
| Omega Psych H | 0.001267 |
| Omega Ordinal | 0.5862 |
| Cronbach Alpha | 0.3679 |
| Greatest Lower Bound | 0.4454 |
| Alpha Ordinal | 0.385 |
Positive correlations: 3 out of 3 (100%)
Scatter matrix
print(x$scatterMatrix$output$scatterMatrix)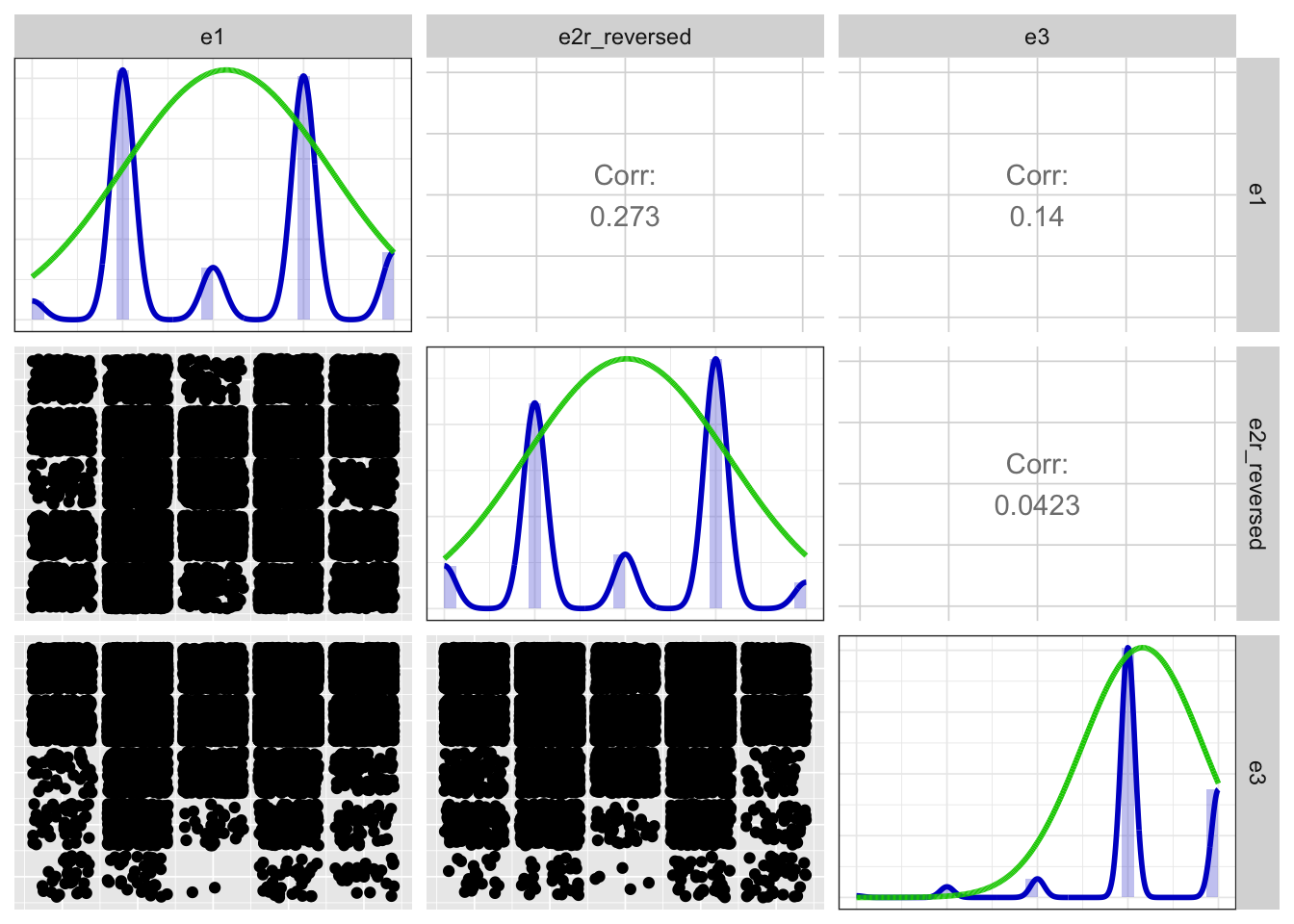
x$scatterMatrix$output$scatterMatrix <- no_md()Detailed output
print(x)##
## Information about this analysis:
##
## Dataframe: res$dat
## Items: e1, e2r_reversed, e3
## Observations: 31446
## Positive correlations: 3 out of 3 (100%)
##
## Estimates assuming interval level:
##
## Omega (total): 0.57
## Omega (hierarchical): 0
## Revelle's omega (total): 0.29
## Greatest Lower Bound (GLB): 0.45
## Coefficient H: 0.91
## Cronbach's alpha: 0.37
## Confidence intervals:
## Omega (total): [0.5, 0.64]
## Cronbach's alpha: [0.36, 0.38]
##
## Estimates assuming ordinal level:
##
## Ordinal Omega (total): 0.59
## Ordinal Omega (hierarch.): 0.59
## Ordinal Cronbach's alpha: 0.38
## Confidence intervals:
## Ordinal Omega (total): [0.52, 0.65]
## Ordinal Cronbach's alpha: [0.37, 0.4]
##
## Note: the normal point estimate and confidence interval for omega are based on the procedure suggested by Dunn, Baguley & Brunsden (2013) using the MBESS function ci.reliability, whereas the psych package point estimate was suggested in Revelle & Zinbarg (2008). See the help ('?scaleStructure') for more information.
##
## Eigen values: 1.326, 0.966, 0.708
## Loadings:
## PC1
## e1 0.789
## e2r_reversed 0.718
## e3 0.433
##
## PC1
## SS loadings 1.326
## Proportion Var 0.442
##
## vars n mean sd median trimmed mad min max range skew kurtosis se
## e1 1 31446 3.15 1.14 3 3.09 1.48 1 5 4 0.02 -1.37 0.01
## e2r_reversed 2 31446 3.02 1.12 3 3.06 1.48 1 5 4 -0.16 -1.30 0.01
## e3 3 31446 4.16 0.67 4 4.22 0.00 1 5 4 -1.09 3.25 0.00Summary statistics
for (i in seq_along(names(items))) {
attributes(items[[i]]) = recursive_escape(attributes(items[[i]]))
}
escaped_table(codebook_table(items))##
##
## name label data_type value_labels n_missing complete_rate min median max mean sd n_value_labels hist
## ------------- -------------------- --------------- ---------------------------------------------------------------------------------------------------------------------------- ---------- -------------- ---- ------- ---- ------ ------- --------------- ---------
## e1 Is talkative haven_labelled 1. Disagree strongly,<br>2. Disagree a little,<br>3. Neither agree nor disagree,<br>4. Agree a little,<br>5. Agree strongly 69795 0.3106 1 3 5 3.146 1.1437 5 ▁▇▁▂▁▇▁▂
## e2r_reversed NA haven_labelled 5. Disagree strongly,<br>4. Disagree a little,<br>3. Neither agree nor disagree,<br>2. Agree a little,<br>1. Agree strongly 69795 0.3106 1 3 5 3.019 1.1246 5 ▂▆▁▂▁▇▁▁
## e3 Outgoing, sociable. haven_labelled 1. Disagree strongly,<br>2. Disagree a little,<br>3. Neither agree nor disagree,<br>4. Agree a little,<br>5. Agree strongly 69795 0.3106 1 4 5 4.162 0.6687 5 ▁▁▁▁▁▇▁▃Scale: big5_con
Overview
Reliability: ωordinal [95% CI] = 0.46 [0.45;0.47].
Missing: 69795.
old_height <- knitr::opts_chunk$get("fig.height")
new_height <- length(scale_info$scale_item_names)
new_height <- ifelse(new_height > 20, 20, new_height)
new_height <- ifelse(new_height < 1, 1, new_height)
new_height <- ifelse(is.na(new_height) | is.nan(new_height),
old_height, new_height)
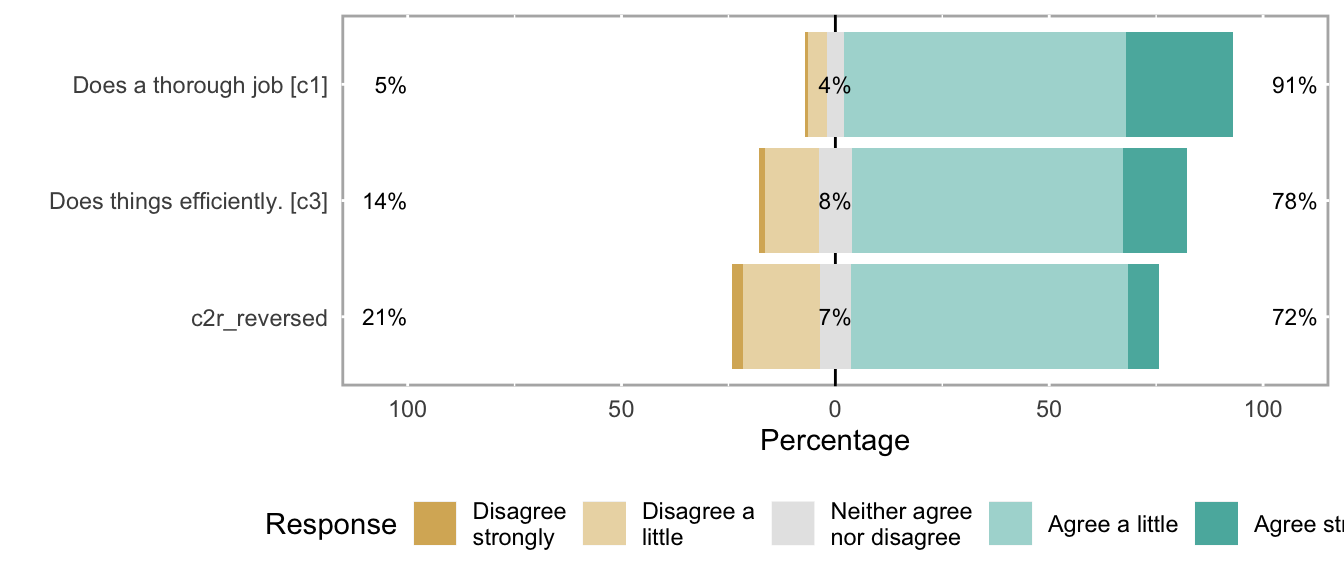
knitr::opts_chunk$set(fig.height = new_height)if (dplyr::n_distinct(na.omit(unlist(items))) < 12) {
likert_plot <- likert_from_items(items)
if (!is.null(likert_plot)) {
graphics::plot(likert_plot)
}
}
knitr::opts_chunk$set(fig.height = old_height)wrap_at <- knitr::opts_chunk$get("fig.width") * 10dist_plot <- plot_labelled(scale, scale_name, wrap_at)
choices <- attributes(items[[1]])$item$choices
breaks <- as.numeric(names(choices))
if (length(breaks)) {
suppressMessages( # ignore message about overwriting x axis
dist_plot <- dist_plot +
ggplot2::scale_x_continuous("values",
breaks = breaks,
labels = stringr::str_wrap(unlist(choices), ceiling(wrap_at * 0.21))) +
ggplot2::expand_limits(x = range(breaks)))
}
dist_plot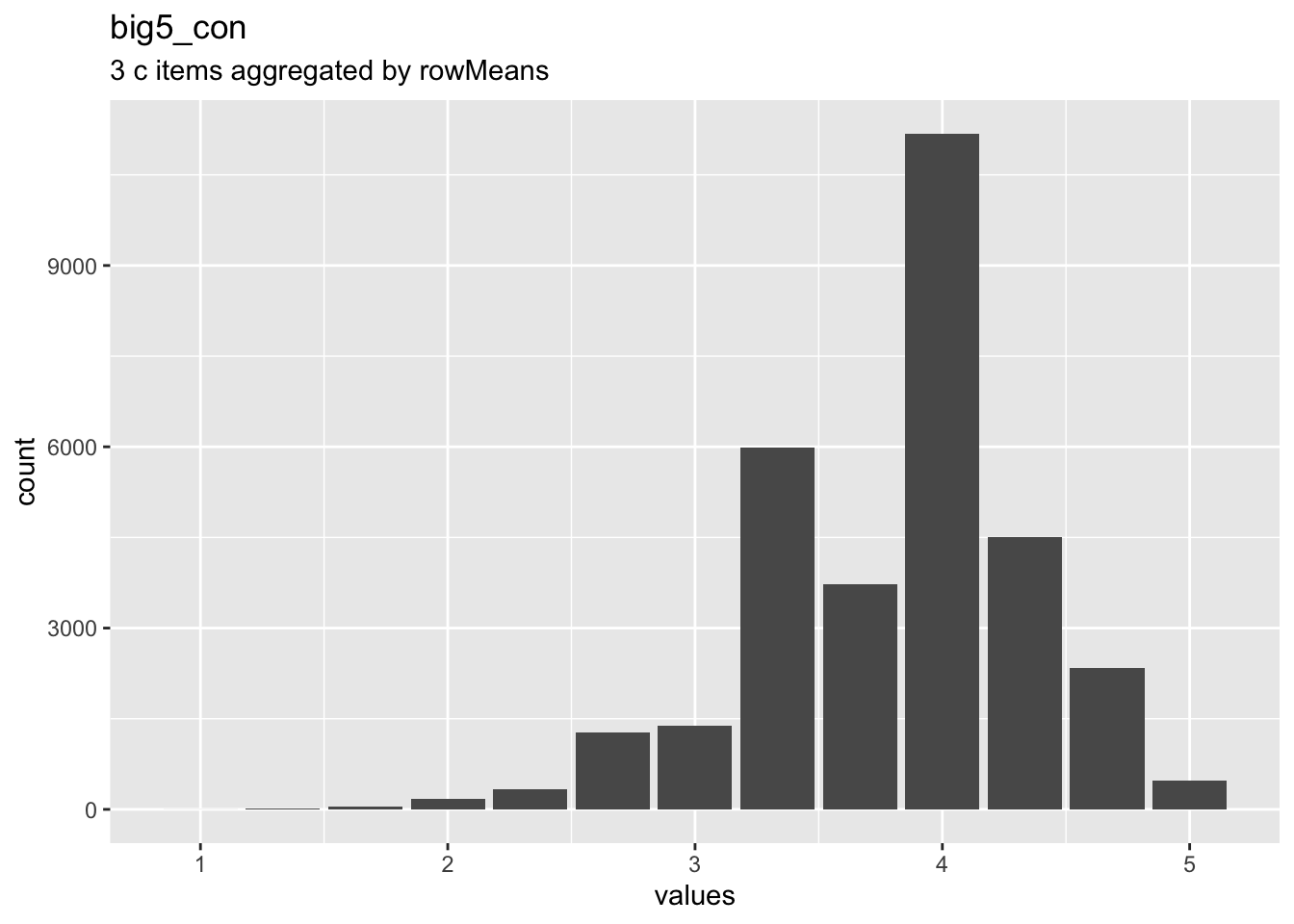
Reliability details
for (i in seq_along(reliabilities)) {
rel <- reliabilities[[i]]
cat(knitr::knit_print(rel, indent = paste0(indent, "####")))
}Reliability Indices
coefs <- x$scaleReliability$output$dat %>%
tidyr::gather(index, estimate) %>%
dplyr::filter(index != "n.items", index != "n.observations") %>%
dplyr::mutate(index = stringr::str_to_title(
stringr::str_replace_all(index,
stringr::fixed("."), " ")))
cis <- coefs %>%
dplyr::filter(stringr::str_detect(index, " Ci ")) %>%
tidyr::separate(index, c("index", "hilo"), sep = " Ci ") %>%
tidyr::spread(hilo, estimate)
if (nrow(cis)) {
cis <- cis %>% dplyr::rename(
`Lower 95% CI` = .data$Lo, `Upper 95% CI` = .data$Hi
)
}
coefs_with_cis <- coefs %>%
dplyr::filter(!stringr::str_detect(index, " Ci ")) %>%
dplyr::left_join(cis, by = "index") %>%
dplyr::mutate(index = dplyr::if_else(index == "Glb", "Greatest Lower Bound", .data$index)) %>%
dplyr::arrange(!stringr::str_detect(index, "Omega")) %>%
dplyr::select(Index = .data$index, Estimate = .data$estimate)
pander::pander(coefs_with_cis)| Index | Estimate |
|---|---|
| Omega | 0.3187 |
| Omega Psych Tot | 0.3566 |
| Omega Psych H | 0.0156 |
| Omega Ordinal | 0.459 |
| Cronbach Alpha | 0.2885 |
| Greatest Lower Bound | 0.3748 |
| Alpha Ordinal | 0.4095 |
Positive correlations: 3 out of 3 (100%)
Scatter matrix
print(x$scatterMatrix$output$scatterMatrix)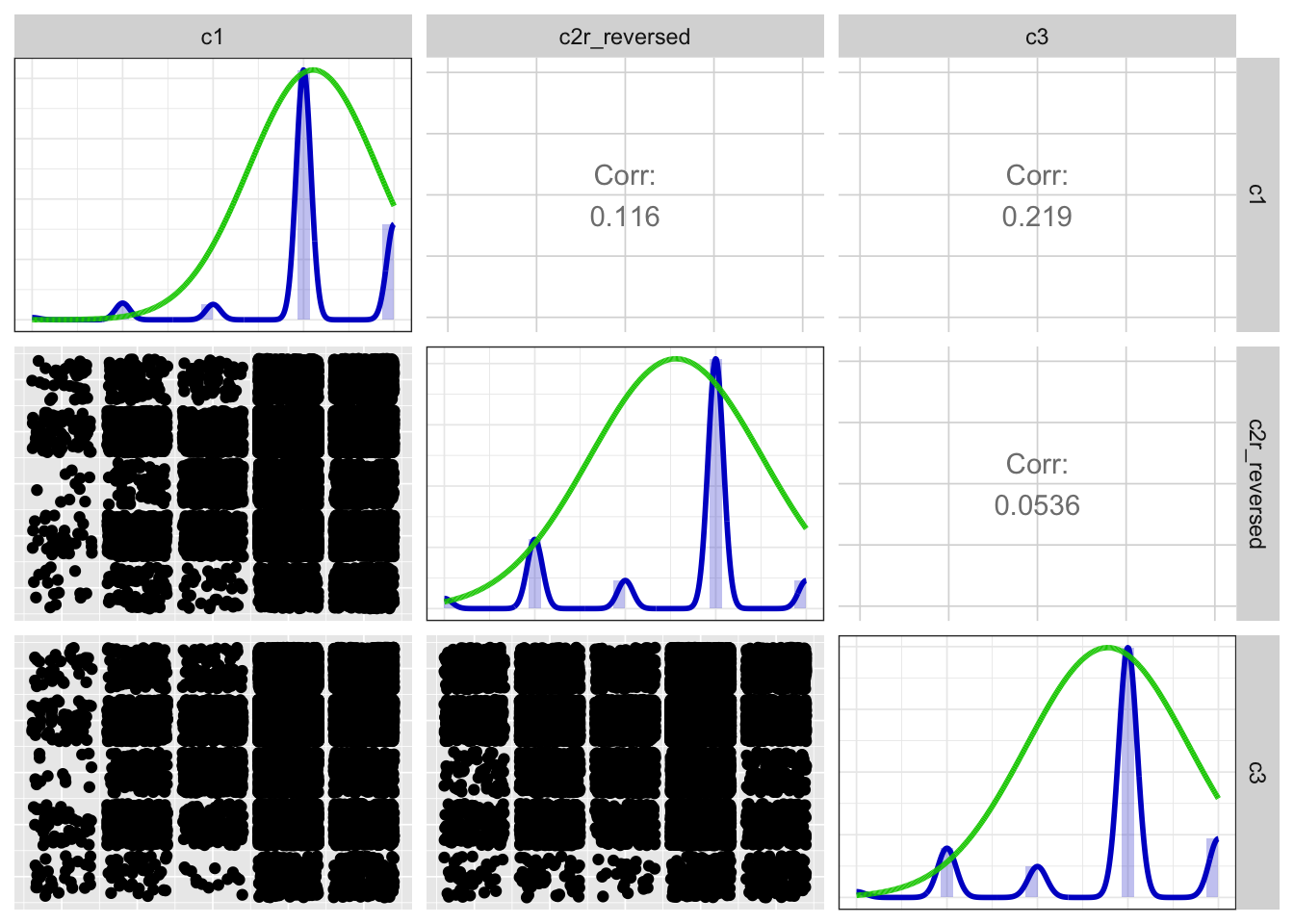
x$scatterMatrix$output$scatterMatrix <- no_md()Detailed output
print(x)##
## Information about this analysis:
##
## Dataframe: res$dat
## Items: c1, c2r_reversed, c3
## Observations: 31446
## Positive correlations: 3 out of 3 (100%)
##
## Estimates assuming interval level:
##
## Omega (total): 0.32
## Omega (hierarchical): 0.02
## Revelle's omega (total): 0.36
## Greatest Lower Bound (GLB): 0.37
## Coefficient H: 0.51
## Cronbach's alpha: 0.29
## Confidence intervals:
## Omega (total): [0.31, 0.33]
## Cronbach's alpha: [0.28, 0.3]
##
## Estimates assuming ordinal level:
##
## Ordinal Omega (total): 0.46
## Ordinal Omega (hierarch.): 0.46
## Ordinal Cronbach's alpha: 0.41
## Confidence intervals:
## Ordinal Omega (total): [0.45, 0.47]
## Ordinal Cronbach's alpha: [0.4, 0.42]
##
## Note: the normal point estimate and confidence interval for omega are based on the procedure suggested by Dunn, Baguley & Brunsden (2013) using the MBESS function ci.reliability, whereas the psych package point estimate was suggested in Revelle & Zinbarg (2008). See the help ('?scaleStructure') for more information.
##
## Eigen values: 1.272, 0.957, 0.771
## Loadings:
## PC1
## c1 0.757
## c2r_reversed 0.459
## c3 0.699
##
## PC1
## SS loadings 1.272
## Proportion Var 0.424
##
## vars n mean sd median trimmed mad min max range skew kurtosis se
## c1 1 31446 4.11 0.71 4 4.19 0 1 5 4 -1.26 3.27 0.00
## c2r_reversed 2 31446 3.56 0.95 4 3.64 0 1 5 4 -1.00 0.08 0.01
## c3 3 31446 3.78 0.90 4 3.86 0 1 5 4 -1.06 0.79 0.01Summary statistics
for (i in seq_along(names(items))) {
attributes(items[[i]]) = recursive_escape(attributes(items[[i]]))
}
escaped_table(codebook_table(items))##
##
## name label data_type value_labels n_missing complete_rate min median max mean sd n_value_labels hist
## ------------- ------------------------- --------------- ---------------------------------------------------------------------------------------------------------------------------- ---------- -------------- ---- ------- ---- ------ ------- --------------- ---------
## c1 Does a thorough job haven_labelled 1. Disagree strongly,<br>2. Disagree a little,<br>3. Neither agree nor disagree,<br>4. Agree a little,<br>5. Agree strongly 69795 0.3106 1 4 5 4.106 0.7131 5 ▁▁▁▁▁▇▁▃
## c2r_reversed NA haven_labelled 5. Disagree strongly,<br>4. Disagree a little,<br>3. Neither agree nor disagree,<br>2. Agree a little,<br>1. Agree strongly 69795 0.3106 1 4 5 3.561 0.9535 5 ▁▂▁▁▁▇▁▁
## c3 Does things efficiently. haven_labelled 1. Disagree strongly,<br>2. Disagree a little,<br>3. Neither agree nor disagree,<br>4. Agree a little,<br>5. Agree strongly 69795 0.3106 1 4 5 3.777 0.8988 5 ▁▂▁▁▁▇▁▂Scale: big5_open
Overview
Reliability: ωordinal [95% CI] = 0.53 [0.52;0.54].
Missing: 69795.
old_height <- knitr::opts_chunk$get("fig.height")
new_height <- length(scale_info$scale_item_names)
new_height <- ifelse(new_height > 20, 20, new_height)
new_height <- ifelse(new_height < 1, 1, new_height)
new_height <- ifelse(is.na(new_height) | is.nan(new_height),
old_height, new_height)
knitr::opts_chunk$set(fig.height = new_height)if (dplyr::n_distinct(na.omit(unlist(items))) < 12) {
likert_plot <- likert_from_items(items)
if (!is.null(likert_plot)) {
graphics::plot(likert_plot)
}
}
knitr::opts_chunk$set(fig.height = old_height)wrap_at <- knitr::opts_chunk$get("fig.width") * 10dist_plot <- plot_labelled(scale, scale_name, wrap_at)
choices <- attributes(items[[1]])$item$choices
breaks <- as.numeric(names(choices))
if (length(breaks)) {
suppressMessages( # ignore message about overwriting x axis
dist_plot <- dist_plot +
ggplot2::scale_x_continuous("values",
breaks = breaks,
labels = stringr::str_wrap(unlist(choices), ceiling(wrap_at * 0.21))) +
ggplot2::expand_limits(x = range(breaks)))
}
dist_plot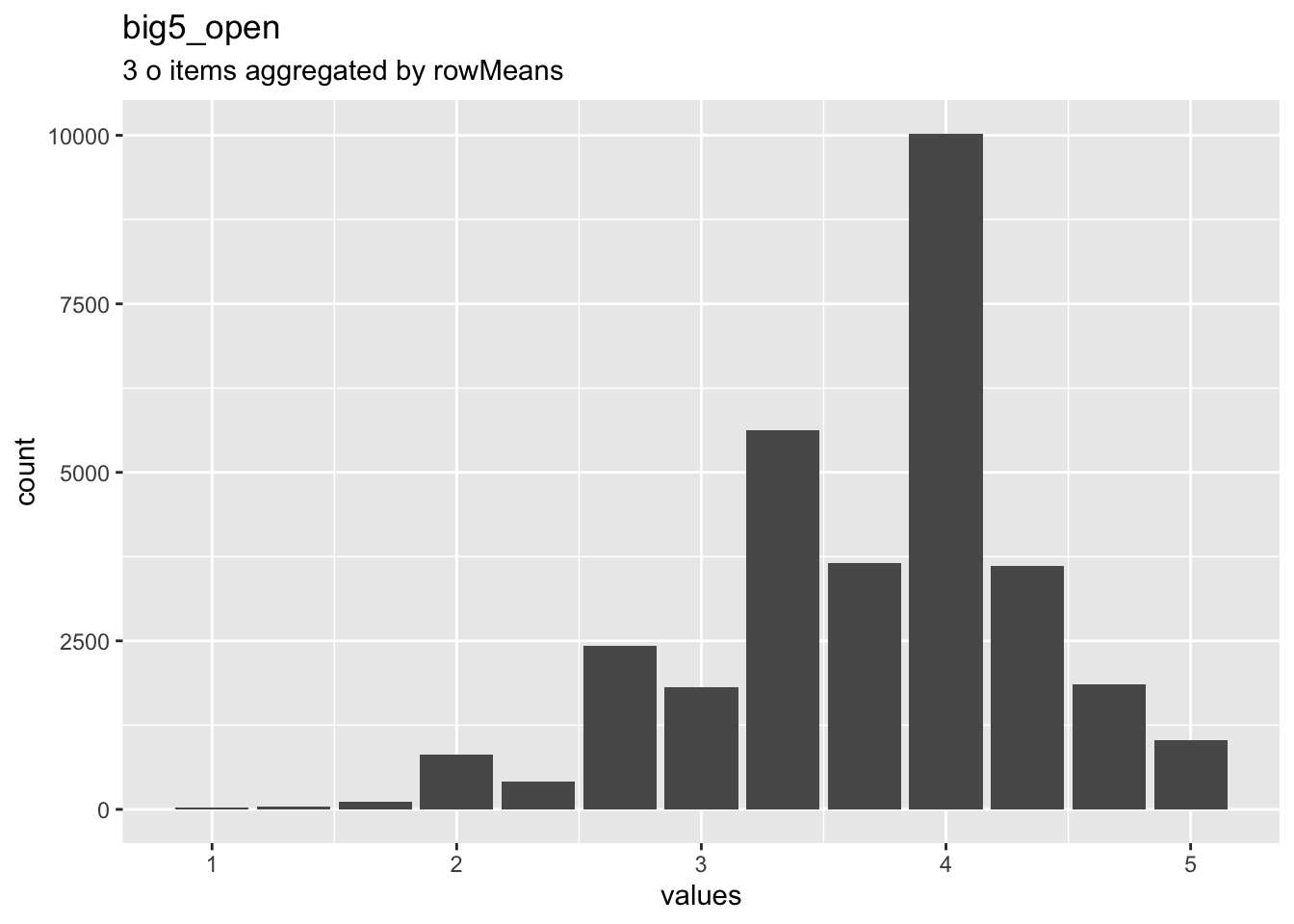
Reliability details
for (i in seq_along(reliabilities)) {
rel <- reliabilities[[i]]
cat(knitr::knit_print(rel, indent = paste0(indent, "####")))
}Reliability Indices
coefs <- x$scaleReliability$output$dat %>%
tidyr::gather(index, estimate) %>%
dplyr::filter(index != "n.items", index != "n.observations") %>%
dplyr::mutate(index = stringr::str_to_title(
stringr::str_replace_all(index,
stringr::fixed("."), " ")))
cis <- coefs %>%
dplyr::filter(stringr::str_detect(index, " Ci ")) %>%
tidyr::separate(index, c("index", "hilo"), sep = " Ci ") %>%
tidyr::spread(hilo, estimate)
if (nrow(cis)) {
cis <- cis %>% dplyr::rename(
`Lower 95% CI` = .data$Lo, `Upper 95% CI` = .data$Hi
)
}
coefs_with_cis <- coefs %>%
dplyr::filter(!stringr::str_detect(index, " Ci ")) %>%
dplyr::left_join(cis, by = "index") %>%
dplyr::mutate(index = dplyr::if_else(index == "Glb", "Greatest Lower Bound", .data$index)) %>%
dplyr::arrange(!stringr::str_detect(index, "Omega")) %>%
dplyr::select(Index = .data$index, Estimate = .data$estimate)
pander::pander(coefs_with_cis)| Index | Estimate |
|---|---|
| Omega | 0.4497 |
| Omega Psych Tot | 0.4581 |
| Omega Psych H | 0.4328 |
| Omega Ordinal | 0.5272 |
| Cronbach Alpha | 0.4453 |
| Greatest Lower Bound | 0.4627 |
| Alpha Ordinal | 0.5229 |
Positive correlations: 3 out of 3 (100%)
Scatter matrix
print(x$scatterMatrix$output$scatterMatrix)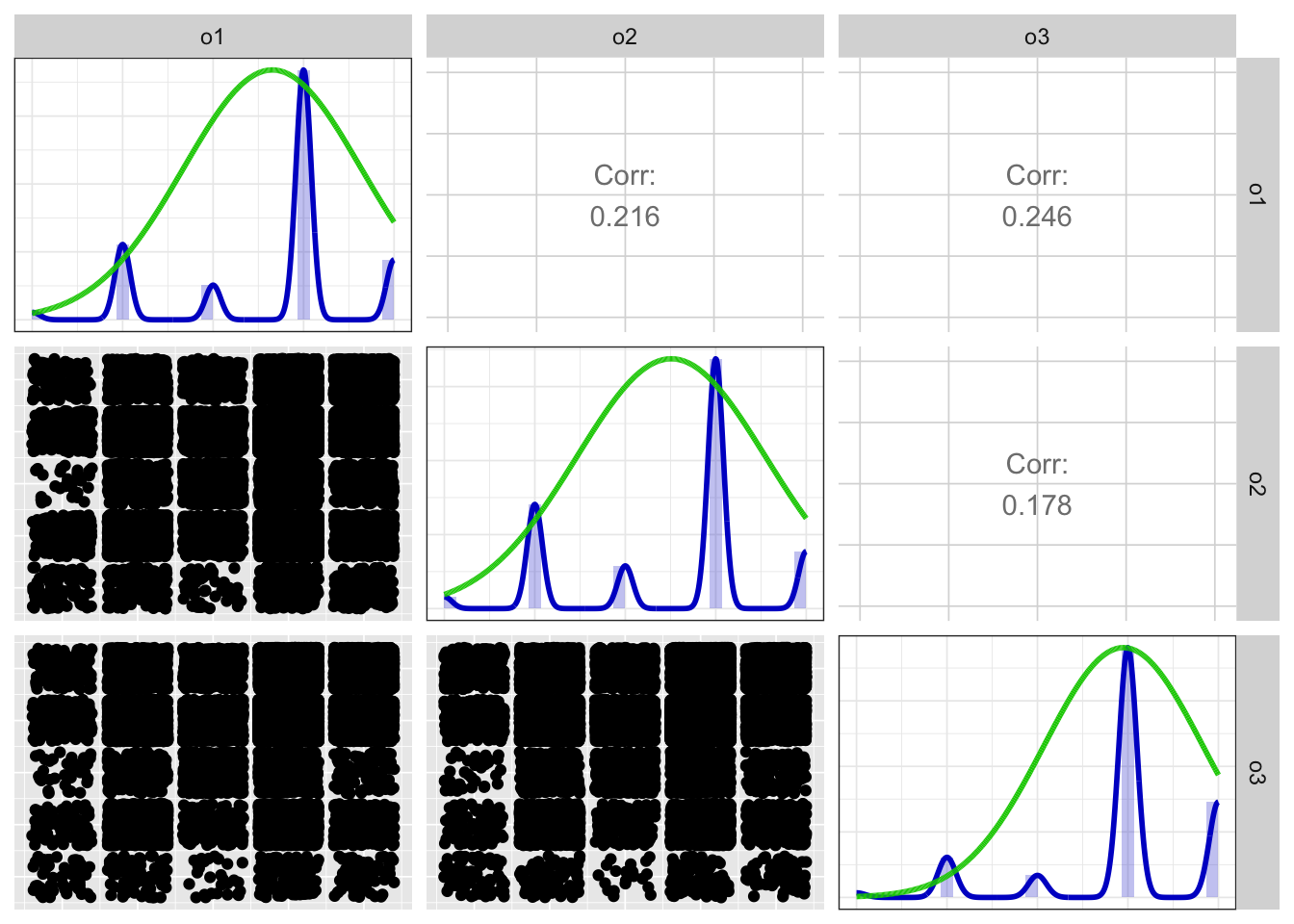
x$scatterMatrix$output$scatterMatrix <- no_md()Detailed output
print(x)##
## Information about this analysis:
##
## Dataframe: res$dat
## Items: o1, o2, o3
## Observations: 31446
## Positive correlations: 3 out of 3 (100%)
##
## Estimates assuming interval level:
##
## Omega (total): 0.45
## Omega (hierarchical): 0.43
## Revelle's omega (total): 0.46
## Greatest Lower Bound (GLB): 0.46
## Coefficient H: 0.46
## Cronbach's alpha: 0.45
## Confidence intervals:
## Omega (total): [0.44, 0.46]
## Cronbach's alpha: [0.43, 0.46]
##
## Estimates assuming ordinal level:
##
## Ordinal Omega (total): 0.53
## Ordinal Omega (hierarch.): 0.53
## Ordinal Cronbach's alpha: 0.52
## Confidence intervals:
## Ordinal Omega (total): [0.52, 0.54]
## Ordinal Cronbach's alpha: [0.51, 0.53]
##
## Note: the normal point estimate and confidence interval for omega are based on the procedure suggested by Dunn, Baguley & Brunsden (2013) using the MBESS function ci.reliability, whereas the psych package point estimate was suggested in Revelle & Zinbarg (2008). See the help ('?scaleStructure') for more information.
##
## Eigen values: 1.428, 0.826, 0.747
## Loadings:
## PC1
## o1 0.726
## o2 0.653
## o3 0.689
##
## PC1
## SS loadings 1.428
## Proportion Var 0.476
##
## vars n mean sd median trimmed mad min max range skew kurtosis se
## o1 1 31446 3.65 0.98 4 3.71 0 1 5 4 -0.83 -0.13 0.01
## o2 2 31446 3.51 1.05 4 3.54 0 1 5 4 -0.62 -0.66 0.01
## o3 3 31446 3.95 0.88 4 4.08 0 1 5 4 -1.16 1.32 0.00Summary statistics
for (i in seq_along(names(items))) {
attributes(items[[i]]) = recursive_escape(attributes(items[[i]]))
}
escaped_table(codebook_table(items))##
##
## name label data_type value_labels n_missing complete_rate min median max mean sd n_value_labels hist
## ----- ---------------------------------------- --------------- ---------------------------------------------------------------------------------------------------------------------------- ---------- -------------- ---- ------- ---- ------ ------- --------------- ---------
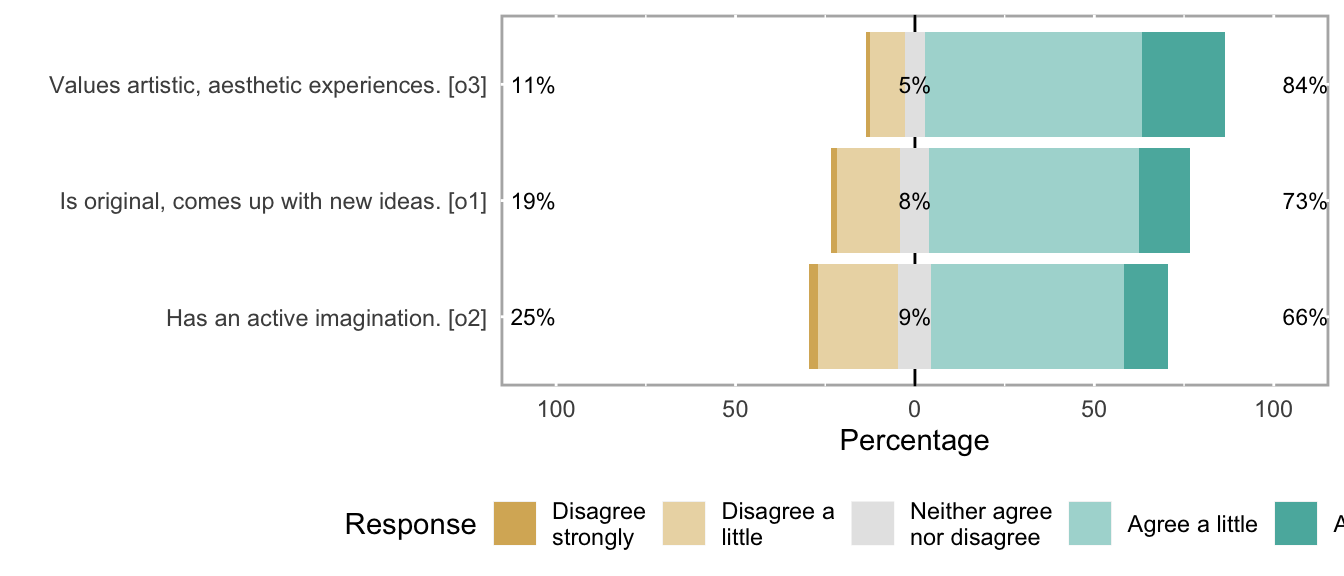
## o1 Is original, comes up with new ideas. haven_labelled 1. Disagree strongly,<br>2. Disagree a little,<br>3. Neither agree nor disagree,<br>4. Agree a little,<br>5. Agree strongly 69795 0.3106 1 4 5 3.654 0.9819 5 ▁▂▁▁▁▇▁▂
## o2 Has an active imagination. haven_labelled 1. Disagree strongly,<br>2. Disagree a little,<br>3. Neither agree nor disagree,<br>4. Agree a little,<br>5. Agree strongly 69795 0.3106 1 4 5 3.508 1.0456 5 ▁▃▁▂▁▇▁▂
## o3 Values artistic, aesthetic experiences. haven_labelled 1. Disagree strongly,<br>2. Disagree a little,<br>3. Neither agree nor disagree,<br>4. Agree a little,<br>5. Agree strongly 69795 0.3106 1 4 5 3.950 0.8793 5 ▁▁▁▁▁▇▁▃Scale: big5_neu
Overview
Reliability: Not computed.
Missing: 69795.
old_height <- knitr::opts_chunk$get("fig.height")
new_height <- length(scale_info$scale_item_names)
new_height <- ifelse(new_height > 20, 20, new_height)
new_height <- ifelse(new_height < 1, 1, new_height)
new_height <- ifelse(is.na(new_height) | is.nan(new_height),
old_height, new_height)
knitr::opts_chunk$set(fig.height = new_height)if (dplyr::n_distinct(na.omit(unlist(items))) < 12) {
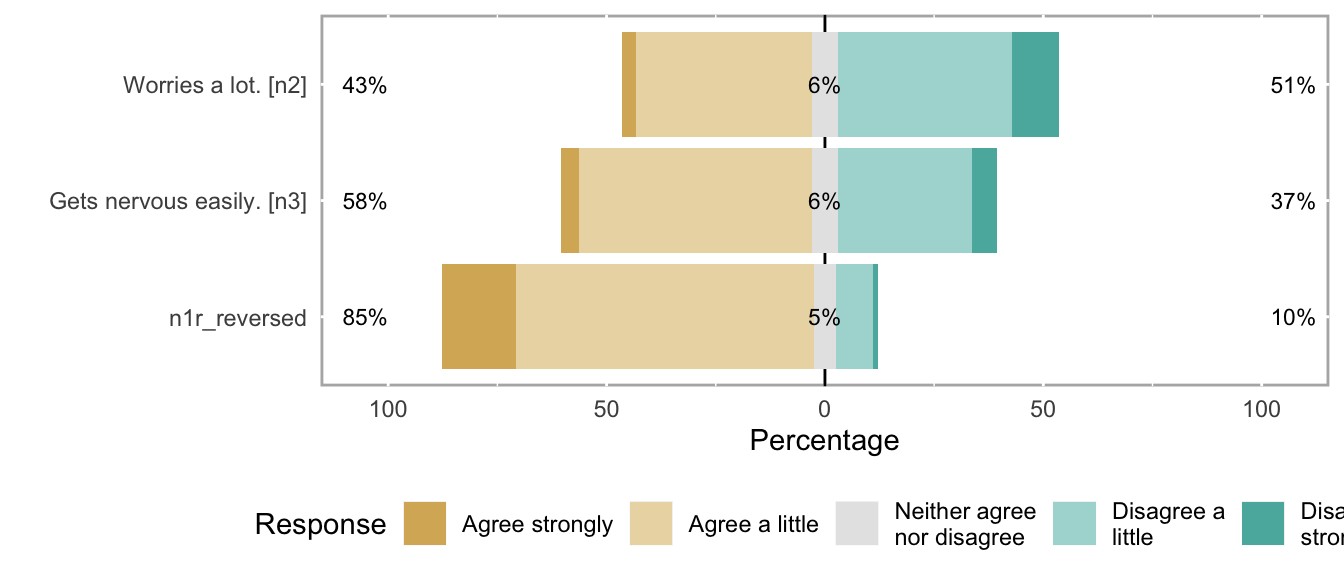
likert_plot <- likert_from_items(items)
if (!is.null(likert_plot)) {
graphics::plot(likert_plot)
}
}
knitr::opts_chunk$set(fig.height = old_height)wrap_at <- knitr::opts_chunk$get("fig.width") * 10dist_plot <- plot_labelled(scale, scale_name, wrap_at)
choices <- attributes(items[[1]])$item$choices
breaks <- as.numeric(names(choices))
if (length(breaks)) {
suppressMessages( # ignore message about overwriting x axis
dist_plot <- dist_plot +
ggplot2::scale_x_continuous("values",
breaks = breaks,
labels = stringr::str_wrap(unlist(choices), ceiling(wrap_at * 0.21))) +
ggplot2::expand_limits(x = range(breaks)))
}
dist_plot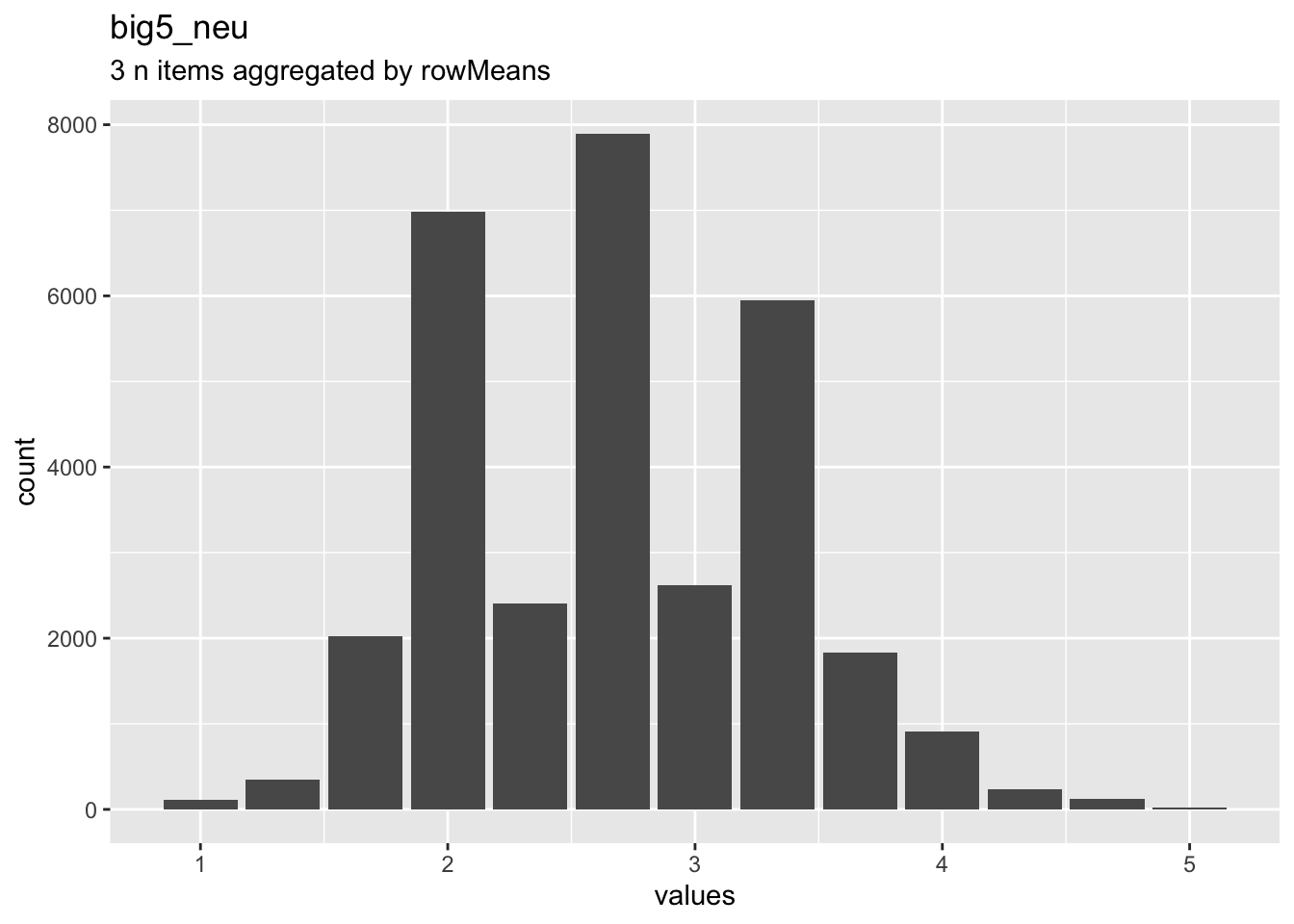
Reliability details
for (i in seq_along(reliabilities)) {
rel <- reliabilities[[i]]
cat(knitr::knit_print(rel, indent = paste0(indent, "####")))
}Summary statistics
for (i in seq_along(names(items))) {
attributes(items[[i]]) = recursive_escape(attributes(items[[i]]))
}
escaped_table(codebook_table(items))##
##
## name label data_type value_labels n_missing complete_rate min median max mean sd n_value_labels hist
## ------------- --------------------- --------------- ---------------------------------------------------------------------------------------------------------------------------- ---------- -------------- ---- ------- ---- ------ ------- --------------- ---------
## n1r_reversed NA haven_labelled 5. Disagree strongly,<br>4. Disagree a little,<br>3. Neither agree nor disagree,<br>2. Agree a little,<br>1. Agree strongly 69795 0.3106 1 2 5 2.087 0.8107 5 ▂▇▁▁▁▁▁▁
## n2 Worries a lot. haven_labelled 1. Disagree strongly,<br>2. Disagree a little,<br>3. Neither agree nor disagree,<br>4. Agree a little,<br>5. Agree strongly 69795 0.3106 1 4 5 3.148 1.1541 5 ▁▇▁▁▁▇▁▂
## n3 Gets nervous easily. haven_labelled 1. Disagree strongly,<br>2. Disagree a little,<br>3. Neither agree nor disagree,<br>4. Agree a little,<br>5. Agree strongly 69795 0.3106 1 2 5 2.806 1.0969 5 ▁▇▁▁▁▅▁▁Scale: big5_agree
Overview
Reliability: ωordinal [95% CI] = 0.57 [0.55;0.59].
Missing: 69795.
old_height <- knitr::opts_chunk$get("fig.height")
new_height <- length(scale_info$scale_item_names)
new_height <- ifelse(new_height > 20, 20, new_height)
new_height <- ifelse(new_height < 1, 1, new_height)
new_height <- ifelse(is.na(new_height) | is.nan(new_height),
old_height, new_height)
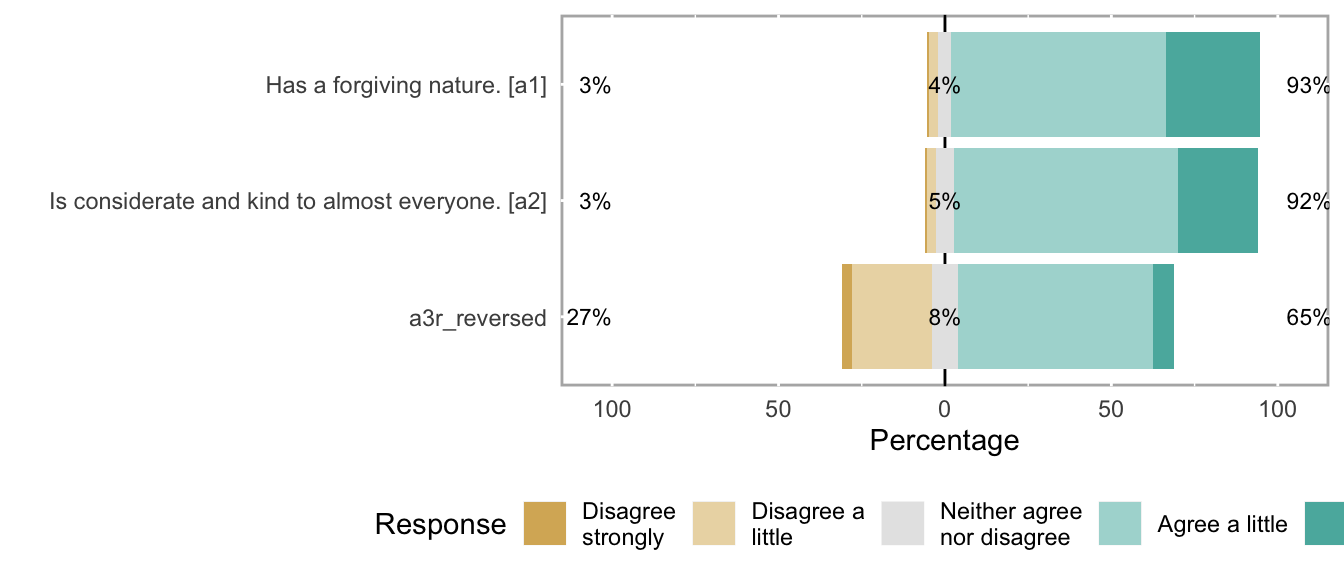
knitr::opts_chunk$set(fig.height = new_height)if (dplyr::n_distinct(na.omit(unlist(items))) < 12) {
likert_plot <- likert_from_items(items)
if (!is.null(likert_plot)) {
graphics::plot(likert_plot)
}
}
knitr::opts_chunk$set(fig.height = old_height)wrap_at <- knitr::opts_chunk$get("fig.width") * 10dist_plot <- plot_labelled(scale, scale_name, wrap_at)
choices <- attributes(items[[1]])$item$choices
breaks <- as.numeric(names(choices))
if (length(breaks)) {
suppressMessages( # ignore message about overwriting x axis
dist_plot <- dist_plot +
ggplot2::scale_x_continuous("values",
breaks = breaks,
labels = stringr::str_wrap(unlist(choices), ceiling(wrap_at * 0.21))) +
ggplot2::expand_limits(x = range(breaks)))
}
dist_plot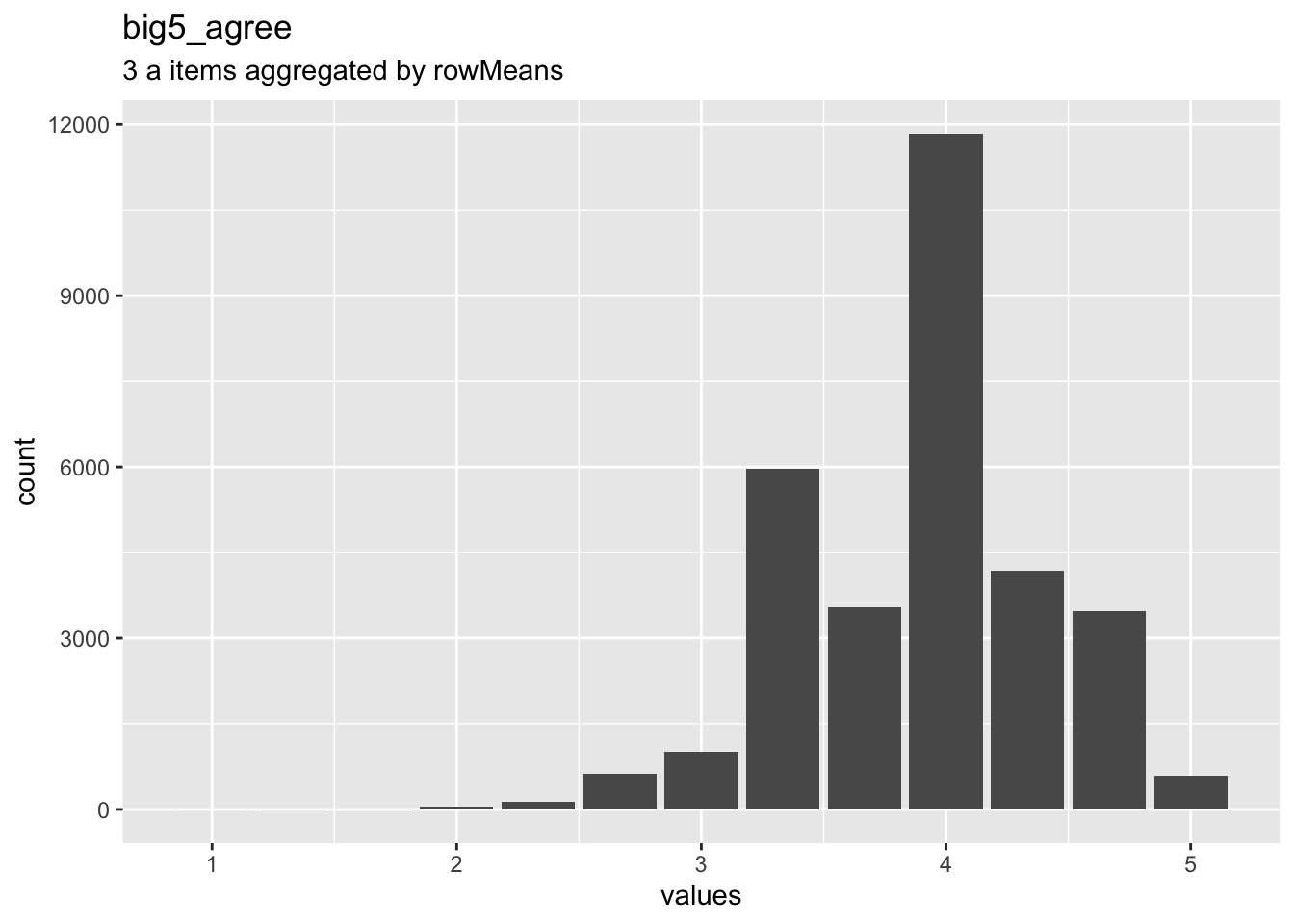
Reliability details
for (i in seq_along(reliabilities)) {
rel <- reliabilities[[i]]
cat(knitr::knit_print(rel, indent = paste0(indent, "####")))
}Reliability Indices
coefs <- x$scaleReliability$output$dat %>%
tidyr::gather(index, estimate) %>%
dplyr::filter(index != "n.items", index != "n.observations") %>%
dplyr::mutate(index = stringr::str_to_title(
stringr::str_replace_all(index,
stringr::fixed("."), " ")))
cis <- coefs %>%
dplyr::filter(stringr::str_detect(index, " Ci ")) %>%
tidyr::separate(index, c("index", "hilo"), sep = " Ci ") %>%
tidyr::spread(hilo, estimate)
if (nrow(cis)) {
cis <- cis %>% dplyr::rename(
`Lower 95% CI` = .data$Lo, `Upper 95% CI` = .data$Hi
)
}
coefs_with_cis <- coefs %>%
dplyr::filter(!stringr::str_detect(index, " Ci ")) %>%
dplyr::left_join(cis, by = "index") %>%
dplyr::mutate(index = dplyr::if_else(index == "Glb", "Greatest Lower Bound", .data$index)) %>%
dplyr::arrange(!stringr::str_detect(index, "Omega")) %>%
dplyr::select(Index = .data$index, Estimate = .data$estimate)
pander::pander(coefs_with_cis)| Index | Estimate |
|---|---|
| Omega | 0.3774 |
| Omega Psych Tot | 0.444 |
| Omega Psych H | 0.01546 |
| Omega Ordinal | 0.5685 |
| Cronbach Alpha | 0.2718 |
| Greatest Lower Bound | 0.4653 |
| Alpha Ordinal | 0.4498 |
Positive correlations: 3 out of 3 (100%)
Scatter matrix
print(x$scatterMatrix$output$scatterMatrix)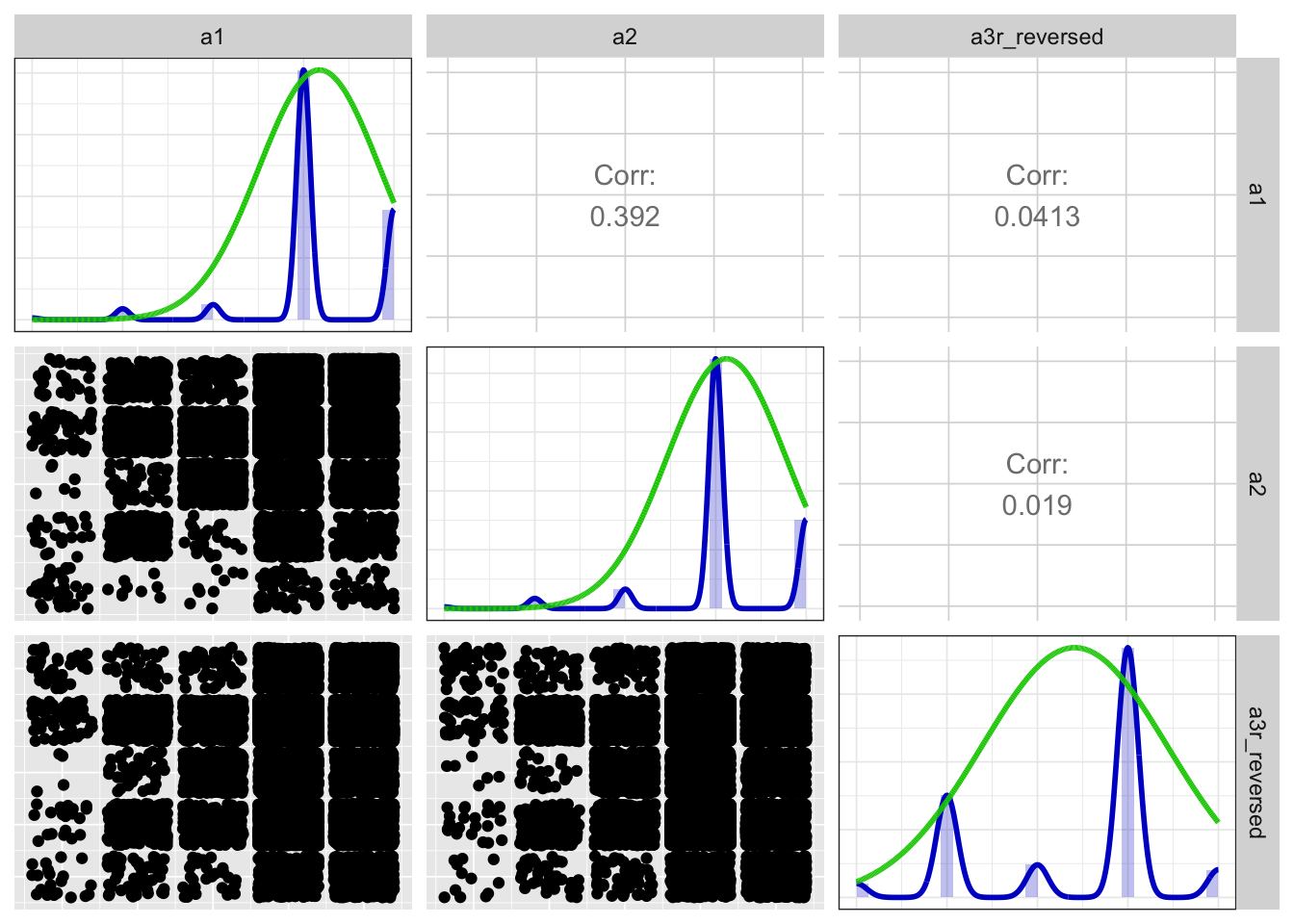
x$scatterMatrix$output$scatterMatrix <- no_md()Detailed output
print(x)##
## Information about this analysis:
##
## Dataframe: res$dat
## Items: a1, a2, a3r_reversed
## Observations: 31446
## Positive correlations: 3 out of 3 (100%)
##
## Estimates assuming interval level:
##
## Omega (total): 0.38
## Omega (hierarchical): 0.02
## Revelle's omega (total): 0.44
## Greatest Lower Bound (GLB): 0.47
## Coefficient H: 0.86
## Cronbach's alpha: 0.27
## Confidence intervals:
## Omega (total): [0.31, 0.44]
## Cronbach's alpha: [0.26, 0.28]
##
## Estimates assuming ordinal level:
##
## Ordinal Omega (total): 0.57
## Ordinal Omega (hierarch.): 0.57
## Ordinal Cronbach's alpha: 0.45
## Confidence intervals:
## Ordinal Omega (total): [0.55, 0.59]
## Ordinal Cronbach's alpha: [0.44, 0.46]
##
## Note: the normal point estimate and confidence interval for omega are based on the procedure suggested by Dunn, Baguley & Brunsden (2013) using the MBESS function ci.reliability, whereas the psych package point estimate was suggested in Revelle & Zinbarg (2008). See the help ('?scaleStructure') for more information.
##
## Eigen values: 1.396, 0.996, 0.608
## Loadings:
## PC1
## a1 0.832
## a2 0.829
## a3r_reversed 0.127
##
## PC1
## SS loadings 1.396
## Proportion Var 0.465
##
## vars n mean sd median trimmed mad min max range skew kurtosis se
## a1 1 31446 4.17 0.67 4 4.23 0 1 5 4 -1.16 3.54 0.00
## a2 2 31446 4.12 0.66 4 4.18 0 1 5 4 -1.11 3.60 0.00
## a3r_reversed 3 31446 3.41 1.02 4 3.47 0 1 5 4 -0.71 -0.68 0.01Summary statistics
for (i in seq_along(names(items))) {
attributes(items[[i]]) = recursive_escape(attributes(items[[i]]))
}
escaped_table(codebook_table(items))##
##
## name label data_type value_labels n_missing complete_rate min median max mean sd n_value_labels hist
## ------------- -------------------------------------------- --------------- ---------------------------------------------------------------------------------------------------------------------------- ---------- -------------- ---- ------- ---- ------ ------- --------------- ---------
## a1 Has a forgiving nature. haven_labelled 1. Disagree strongly,<br>2. Disagree a little,<br>3. Neither agree nor disagree,<br>4. Agree a little,<br>5. Agree strongly 69795 0.3106 1 4 5 4.175 0.6696 5 ▁▁▁▁▁▇▁▃
## a2 Is considerate and kind to almost everyone. haven_labelled 1. Disagree strongly,<br>2. Disagree a little,<br>3. Neither agree nor disagree,<br>4. Agree a little,<br>5. Agree strongly 69795 0.3106 1 4 5 4.119 0.6567 5 ▁▁▁▁▁▇▁▃
## a3r_reversed NA haven_labelled 5. Disagree strongly,<br>4. Disagree a little,<br>3. Neither agree nor disagree,<br>2. Agree a little,<br>1. Agree strongly 69795 0.3106 1 4 5 3.412 1.0235 5 ▁▃▁▁▁▇▁▁missingness_reportitemsCodebook table
export_table(metadata_table)jsonldJSON-LD metadata
The following JSON-LD can be found by search engines, if you share this codebook publicly on the web.
{
"name": "Indonesian Family Life Study, merged subset",
"description": "Data from the IFLS, merged across waves, most outcomes taken from wave 5. Includes birth order, family structure, Big 5 Personality, intelligence tests, and risk lotteries\n\n\n## Table of variables\nThis table contains variable names, labels, and number of missing values.\nSee the complete codebook for more.\n\n[truncated]\n\n### Note\nThis dataset was automatically described using the [codebook R package](https://rubenarslan.github.io/codebook/) (version 0.8.2).",
"identifier": "https://www.rand.org/well-being/social-and-behavioral-policy/data/FLS/IFLS.html",
"creator": "RAND corporation",
"citation": "Strauss, J., Witoelar, F., & Sikoki, B. (2016). The Fifth Wave of the Indonesia Family Life Survey: Overview and Field Report. WR-1143/1-NIA/NICHD",
"url": "https://www.rand.org/well-being/social-and-behavioral-policy/data/FLS/IFLS.html",
"datePublished": "2016",
"temporalCoverage": "2014/2015",
"spatialCoverage": "13 Indonesian provinces. The sample is representative of about 83% of the Indonesian population and contains over 30,000 individuals living in 13 of the 27 provinces in the country. See URL for more.",
"keywords": ["wave", "mother_pidlink", "chron_order_birth", "lifebirths", "multiple_birth", "alive", "birthdate", "any_multiple_birth", "marriage_id", "birthorder_uterus_preg", "sibling_count_uterus_preg", "birthorder_uterus_alive", "sibling_count_uterus_alive", "birthorder_genes", "sibling_count_genes", "pidlink", "father_pidlink", "age", "death_yr", "death_month", "sc05", "province", "sc01_14_14", "sibling_count_naive_ind", "any_multiple_birthdate", "birthorder_naive", "sibling_count_naive", "age_2015_old", "age_2015_young", "raven_2015_young", "math_2015_young", "raven_2015_old", "math_2015_old", "words_immediate", "words_delayed", "words_remembered_avg", "adaptive_numbering", "age_2007_young", "age_2007_old", "raven_2007_old", "raven_2007_young", "math_2007_young", "math_2007_old", "count_backwards", "g_factor_2015_old", "g_factor_2015_young", "g_factor_2007_old", "g_factor_2007_young", "e1", "c1", "o1", "e2r", "n1r", "a1", "n2", "o2", "c2r", "o3", "a2", "c3", "e3", "a3r", "n3", "e2r_reversed", "big5_ext", "c2r_reversed", "big5_con", "big5_open", "n1r_reversed", "big5_neu", "a3r_reversed", "big5_agree", "random_si", "si01", "si02", "si03", "si04", "si05", "si11", "si12", "si13", "si14", "si15", "riskA", "riskB", "attended_school", "highest_education", "currently_attending_school", "hours_in_class", "years_of_education", "Type_of_test_elementary", "Indonesia_score_elementary", "English_score_elementary", "Math_score_elemenatry", "Total_score_elemenatry", "Type_of_test_Junior_High", "Indonesia_score_Junior_High", "English_score_Junior_High", "Math_score_Junior_High", "Total_score_Junior_High", "Type_of_test_Senior_High", "Indonesia_score_Senior_High", "English_score_Senior_High", "Math_score_Senior_High", "Total_score_Senior_High", "Total_score_highest", "Total_score_highest_type", "Math_score_highest", "Math_score_highest_type", "Elementary_worked", "Junior_high_worked", "Senior_high_worked", "University_worked", "total_worked", "Elementary_missed", "Junior_high_missed", "Senior_high_missed", "University_missed", "total_missed", "Category", "Sector", "Self_employed", "ever_smoked", "still_smoking", "amount", "age_first_smoke", "amount_still_smokers", "male", "wage_last_month_log", "wage_last_year_log", "money_spent_smoking_log", "birthyear", "birthorder_naive_factor", "sibling_count_naive_factor", "birthorder_uterus_alive_factor", "sibling_count_uterus_alive_factor", "birthorder_uterus_preg_factor", "sibling_count_uterus_preg_factor", "birthorder_genes_factor", "sibling_count_genes_factor", "count_birthorder_naive", "count_birthorder_uterus_alive", "count_birthorder_uterus_preg", "count_birthorder_genes", "sibling_count", "birth_order_nonlinear", "birth_order", "count_birth_order"],
"@context": "http://schema.org/",
"@type": "Dataset",
"variableMeasured": [
{
"name": "wave",
"@type": "propertyValue"
},
{
"name": "mother_pidlink",
"@type": "propertyValue"
},
{
"name": "chron_order_birth",
"description": "Chronological order of pregnancy's outcome",
"@type": "propertyValue"
},
{
"name": "lifebirths",
"value": "1. 2,\n2. 3,\n3. 4",
"@type": "propertyValue"
},
{
"name": "multiple_birth",
"@type": "propertyValue"
},
{
"name": "alive",
"@type": "propertyValue"
},
{
"name": "birthdate",
"@type": "propertyValue"
},
{
"name": "any_multiple_birth",
"@type": "propertyValue"
},
{
"name": "marriage_id",
"@type": "propertyValue"
},
{
"name": "birthorder_uterus_preg",
"@type": "propertyValue"
},
{
"name": "sibling_count_uterus_preg",
"@type": "propertyValue"
},
{
"name": "birthorder_uterus_alive",
"@type": "propertyValue"
},
{
"name": "sibling_count_uterus_alive",
"@type": "propertyValue"
},
{
"name": "birthorder_genes",
"@type": "propertyValue"
},
{
"name": "sibling_count_genes",
"@type": "propertyValue"
},
{
"name": "pidlink",
"@type": "propertyValue"
},
{
"name": "father_pidlink",
"@type": "propertyValue"
},
{
"name": "age",
"description": "Age now",
"@type": "propertyValue"
},
{
"name": "death_yr",
"@type": "propertyValue"
},
{
"name": "death_month",
"description": "Departure/entry into HH (Month)",
"@type": "propertyValue"
},
{
"name": "sc05",
"description": "Urban/Rural",
"value": "1. 1:Urban,\n2. 2:Rural",
"maxValue": 2,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "province",
"@type": "propertyValue"
},
{
"name": "sc01_14_14",
"description": "2014 Province, 2014 BPS Code",
"@type": "propertyValue"
},
{
"name": "sibling_count_naive_ind",
"@type": "propertyValue"
},
{
"name": "any_multiple_birthdate",
"@type": "propertyValue"
},
{
"name": "birthorder_naive",
"@type": "propertyValue"
},
{
"name": "sibling_count_naive",
"@type": "propertyValue"
},
{
"name": "age_2015_old",
"@type": "propertyValue"
},
{
"name": "age_2015_young",
"@type": "propertyValue"
},
{
"name": "raven_2015_young",
"@type": "propertyValue"
},
{
"name": "math_2015_young",
"@type": "propertyValue"
},
{
"name": "raven_2015_old",
"@type": "propertyValue"
},
{
"name": "math_2015_old",
"@type": "propertyValue"
},
{
"name": "words_immediate",
"description": "count of immediate recall words",
"@type": "propertyValue"
},
{
"name": "words_delayed",
"description": "count of delayed recall words",
"@type": "propertyValue"
},
{
"name": "words_remembered_avg",
"@type": "propertyValue"
},
{
"name": "adaptive_numbering",
"description": "W-score",
"@type": "propertyValue"
},
{
"name": "age_2007_young",
"@type": "propertyValue"
},
{
"name": "age_2007_old",
"@type": "propertyValue"
},
{
"name": "raven_2007_old",
"@type": "propertyValue"
},
{
"name": "raven_2007_young",
"@type": "propertyValue"
},
{
"name": "math_2007_young",
"@type": "propertyValue"
},
{
"name": "math_2007_old",
"@type": "propertyValue"
},
{
"name": "count_backwards",
"@type": "propertyValue"
},
{
"name": "g_factor_2015_old",
"@type": "propertyValue"
},
{
"name": "g_factor_2015_young",
"@type": "propertyValue"
},
{
"name": "g_factor_2007_old",
"@type": "propertyValue"
},
{
"name": "g_factor_2007_young",
"@type": "propertyValue"
},
{
"name": "e1",
"description": "Is talkative",
"value": "1. Disagree strongly,\n2. Disagree a little,\n3. Neither agree nor disagree,\n4. Agree a little,\n5. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "c1",
"description": "Does a thorough job",
"value": "1. Disagree strongly,\n2. Disagree a little,\n3. Neither agree nor disagree,\n4. Agree a little,\n5. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "o1",
"description": "Is original, comes up with new ideas.",
"value": "1. Disagree strongly,\n2. Disagree a little,\n3. Neither agree nor disagree,\n4. Agree a little,\n5. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "e2r",
"value": "1. Disagree strongly,\n2. Disagree a little,\n3. Neither agree nor disagree,\n4. Agree a little,\n5. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "n1r",
"value": "1. Disagree strongly,\n2. Disagree a little,\n3. Neither agree nor disagree,\n4. Agree a little,\n5. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "a1",
"description": "Has a forgiving nature.",
"value": "1. Disagree strongly,\n2. Disagree a little,\n3. Neither agree nor disagree,\n4. Agree a little,\n5. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "n2",
"description": "Worries a lot.",
"value": "1. Disagree strongly,\n2. Disagree a little,\n3. Neither agree nor disagree,\n4. Agree a little,\n5. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "o2",
"description": "Has an active imagination.",
"value": "1. Disagree strongly,\n2. Disagree a little,\n3. Neither agree nor disagree,\n4. Agree a little,\n5. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "c2r",
"value": "1. Disagree strongly,\n2. Disagree a little,\n3. Neither agree nor disagree,\n4. Agree a little,\n5. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "o3",
"description": "Values artistic, aesthetic experiences.",
"value": "1. Disagree strongly,\n2. Disagree a little,\n3. Neither agree nor disagree,\n4. Agree a little,\n5. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "a2",
"description": "Is considerate and kind to almost everyone.",
"value": "1. Disagree strongly,\n2. Disagree a little,\n3. Neither agree nor disagree,\n4. Agree a little,\n5. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "c3",
"description": "Does things efficiently.",
"value": "1. Disagree strongly,\n2. Disagree a little,\n3. Neither agree nor disagree,\n4. Agree a little,\n5. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "e3",
"description": "Outgoing, sociable.",
"value": "1. Disagree strongly,\n2. Disagree a little,\n3. Neither agree nor disagree,\n4. Agree a little,\n5. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "a3r",
"value": "1. Disagree strongly,\n2. Disagree a little,\n3. Neither agree nor disagree,\n4. Agree a little,\n5. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "n3",
"description": "Gets nervous easily.",
"value": "1. Disagree strongly,\n2. Disagree a little,\n3. Neither agree nor disagree,\n4. Agree a little,\n5. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "e2r_reversed",
"value": "5. Disagree strongly,\n4. Disagree a little,\n3. Neither agree nor disagree,\n2. Agree a little,\n1. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "big5_ext",
"description": "3 e items aggregated by rowMeans",
"@type": "propertyValue"
},
{
"name": "c2r_reversed",
"value": "5. Disagree strongly,\n4. Disagree a little,\n3. Neither agree nor disagree,\n2. Agree a little,\n1. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "big5_con",
"description": "3 c items aggregated by rowMeans",
"@type": "propertyValue"
},
{
"name": "big5_open",
"description": "3 o items aggregated by rowMeans",
"@type": "propertyValue"
},
{
"name": "n1r_reversed",
"value": "5. Disagree strongly,\n4. Disagree a little,\n3. Neither agree nor disagree,\n2. Agree a little,\n1. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "big5_neu",
"description": "3 n items aggregated by rowMeans",
"@type": "propertyValue"
},
{
"name": "a3r_reversed",
"value": "5. Disagree strongly,\n4. Disagree a little,\n3. Neither agree nor disagree,\n2. Agree a little,\n1. Agree strongly",
"maxValue": 5,
"minValue": 1,
"@type": "propertyValue"
},
{
"name": "big5_agree",
"description": "3 a items aggregated by rowMeans",
"@type": "propertyValue"
},
{
"name": "random_si",
"value": "1. 1,\n2. 2",
"@type": "propertyValue"
},
{
"name": "si01",
"@type": "propertyValue"
},
{
"name": "si02",
"@type": "propertyValue"
},
{
"name": "si03",
"@type": "propertyValue"
},
{
"name": "si04",
"@type": "propertyValue"
},
{
"name": "si05",
"@type": "propertyValue"
},
{
"name": "si11",
"@type": "propertyValue"
},
{
"name": "si12",
"@type": "propertyValue"
},
{
"name": "si13",
"@type": "propertyValue"
},
{
"name": "si14",
"@type": "propertyValue"
},
{
"name": "si15",
"@type": "propertyValue"
},
{
"name": "riskA",
"@type": "propertyValue"
},
{
"name": "riskB",
"@type": "propertyValue"
},
{
"name": "attended_school",
"@type": "propertyValue"
},
{
"name": "highest_education",
"value": "1. Elementary,\n2. Junior High,\n3. Senior High,\n4. University",
"@type": "propertyValue"
},
{
"name": "currently_attending_school",
"value": "1. no,\n2. yes",
"@type": "propertyValue"
},
{
"name": "hours_in_class",
"@type": "propertyValue"
},
{
"name": "years_of_education",
"@type": "propertyValue"
},
{
"name": "Type_of_test_elementary",
"value": "1. EBTANAS,\n2. UAN/UN",
"@type": "propertyValue"
},
{
"name": "Indonesia_score_elementary",
"@type": "propertyValue"
},
{
"name": "English_score_elementary",
"@type": "propertyValue"
},
{
"name": "Math_score_elemenatry",
"@type": "propertyValue"
},
{
"name": "Total_score_elemenatry",
"description": "Total EBTANAS/UAN/UN score",
"@type": "propertyValue"
},
{
"name": "Type_of_test_Junior_High",
"value": "1. EBTANAS,\n2. UAN/UN",
"@type": "propertyValue"
},
{
"name": "Indonesia_score_Junior_High",
"@type": "propertyValue"
},
{
"name": "English_score_Junior_High",
"@type": "propertyValue"
},
{
"name": "Math_score_Junior_High",
"@type": "propertyValue"
},
{
"name": "Total_score_Junior_High",
"description": "Total EBTANAS/UAN/UN score",
"@type": "propertyValue"
},
{
"name": "Type_of_test_Senior_High",
"value": "1. EBTANAS,\n2. UAN/UN",
"@type": "propertyValue"
},
{
"name": "Indonesia_score_Senior_High",
"@type": "propertyValue"
},
{
"name": "English_score_Senior_High",
"@type": "propertyValue"
},
{
"name": "Math_score_Senior_High",
"@type": "propertyValue"
},
{
"name": "Total_score_Senior_High",
"description": "Total EBTANAS/UAN/UN score",
"@type": "propertyValue"
},
{
"name": "Total_score_highest",
"@type": "propertyValue"
},
{
"name": "Total_score_highest_type",
"value": "1. Elementary,\n2. Junior High,\n3. Senior High",
"@type": "propertyValue"
},
{
"name": "Math_score_highest",
"@type": "propertyValue"
},
{
"name": "Math_score_highest_type",
"value": "1. Elementary,\n2. Junior High,\n3. Senior High",
"@type": "propertyValue"
},
{
"name": "Elementary_worked",
"@type": "propertyValue"
},
{
"name": "Junior_high_worked",
"@type": "propertyValue"
},
{
"name": "Senior_high_worked",
"@type": "propertyValue"
},
{
"name": "University_worked",
"@type": "propertyValue"
},
{
"name": "total_worked",
"@type": "propertyValue"
},
{
"name": "Elementary_missed",
"@type": "propertyValue"
},
{
"name": "Junior_high_missed",
"@type": "propertyValue"
},
{
"name": "Senior_high_missed",
"@type": "propertyValue"
},
{
"name": "University_missed",
"@type": "propertyValue"
},
{
"name": "total_missed",
"@type": "propertyValue"
},
{
"name": "Category",
"value": "1. Casual worker in agriculture,\n2. Casual worker not in agriculture,\n3. Government worker,\n4. Private worker,\n5. Self-employed,\n6. Unpaid family worker",
"@type": "propertyValue"
},
{
"name": "Sector",
"value": "1. Agriculture, forestry, fishing and hunting,\n2. Construction,\n3. Electricity, gas, water,\n4. Finance, insurance, real estate and business services,\n5. Manufacturing,\n6. Mining and quarrying,\n7. Social services,\n8. Transportation, storage and communications,\n9. Wholesale, retail, restaurants and hotels",
"@type": "propertyValue"
},
{
"name": "Self_employed",
"@type": "propertyValue"
},
{
"name": "ever_smoked",
"@type": "propertyValue"
},
{
"name": "still_smoking",
"@type": "propertyValue"
},
{
"name": "amount",
"@type": "propertyValue"
},
{
"name": "age_first_smoke",
"description": "At what age did you start to smoke on a regular basis?",
"@type": "propertyValue"
},
{
"name": "amount_still_smokers",
"@type": "propertyValue"
},
{
"name": "male",
"@type": "propertyValue"
},
{
"name": "wage_last_month_log",
"@type": "propertyValue"
},
{
"name": "wage_last_year_log",
"@type": "propertyValue"
},
{
"name": "money_spent_smoking_log",
"@type": "propertyValue"
},
{
"name": "birthyear",
"@type": "propertyValue"
},
{
"name": "birthorder_naive_factor",
"value": "1. 1,\n2. 2,\n3. 3,\n4. 4,\n5. 5,\n6. >5",
"@type": "propertyValue"
},
{
"name": "sibling_count_naive_factor",
"value": "1. 2,\n2. 3,\n3. 4,\n4. 5,\n5. >5",
"@type": "propertyValue"
},
{
"name": "birthorder_uterus_alive_factor",
"value": "1. 1,\n2. 2,\n3. 3,\n4. 4,\n5. 5,\n6. >5",
"@type": "propertyValue"
},
{
"name": "sibling_count_uterus_alive_factor",
"value": "1. 2,\n2. 3,\n3. 4,\n4. 5,\n5. >5",
"@type": "propertyValue"
},
{
"name": "birthorder_uterus_preg_factor",
"value": "1. 1,\n2. 2,\n3. 3,\n4. 4,\n5. 5,\n6. >5",
"@type": "propertyValue"
},
{
"name": "sibling_count_uterus_preg_factor",
"value": "1. 2,\n2. 3,\n3. 4,\n4. 5,\n5. >5",
"@type": "propertyValue"
},
{
"name": "birthorder_genes_factor",
"value": "1. 1,\n2. 2,\n3. 3,\n4. 4,\n5. 5,\n6. >5",
"@type": "propertyValue"
},
{
"name": "sibling_count_genes_factor",
"value": "1. 2,\n2. 3,\n3. 4,\n4. 5,\n5. >5",
"@type": "propertyValue"
},
{
"name": "count_birthorder_naive",
"value": "1. 1/2,\n2. 2/2,\n3. 1/3,\n4. 2/3,\n5. 3/3,\n6. 1/4,\n7. 2/4,\n8. 3/4,\n9. 4/4,\n10. 1/5,\n11. 2/5,\n12. 3/5,\n13. 4/5,\n14. 5/5,\n15. 1/>5,\n16. 2/>5,\n17. 3/>5,\n18. 4/>5,\n19. 5/>5,\n20. >5/>5",
"@type": "propertyValue"
},
{
"name": "count_birthorder_uterus_alive",
"value": "1. 1/2,\n2. 2/2,\n3. 1/3,\n4. 2/3,\n5. 3/3,\n6. 1/4,\n7. 2/4,\n8. 3/4,\n9. 4/4,\n10. 1/5,\n11. 2/5,\n12. 3/5,\n13. 4/5,\n14. 5/5,\n15. 1/>5,\n16. 2/>5,\n17. 3/>5,\n18. 4/>5,\n19. 5/>5,\n20. >5/>5",
"@type": "propertyValue"
},
{
"name": "count_birthorder_uterus_preg",
"value": "1. 1/2,\n2. 2/2,\n3. 1/3,\n4. 2/3,\n5. 3/3,\n6. 1/4,\n7. 2/4,\n8. 3/4,\n9. 4/4,\n10. 1/5,\n11. 2/5,\n12. 3/5,\n13. 4/5,\n14. 5/5,\n15. 1/>5,\n16. 2/>5,\n17. 3/>5,\n18. 4/>5,\n19. 5/>5,\n20. >5/>5",
"@type": "propertyValue"
},
{
"name": "count_birthorder_genes",
"value": "1. 1/2,\n2. 2/2,\n3. 1/3,\n4. 2/3,\n5. 3/3,\n6. 1/4,\n7. 2/4,\n8. 3/4,\n9. 4/4,\n10. 1/5,\n11. 2/5,\n12. 3/5,\n13. 4/5,\n14. 5/5,\n15. 1/>5,\n16. 2/>5,\n17. 3/>5,\n18. 4/>5,\n19. 5/>5,\n20. >5/>5",
"@type": "propertyValue"
},
{
"name": "sibling_count",
"value": "1. 2,\n2. 3,\n3. 4,\n4. 5,\n5. >5",
"@type": "propertyValue"
},
{
"name": "birth_order_nonlinear",
"value": "1. 1,\n2. 2,\n3. 3,\n4. 4,\n5. 5,\n6. >5",
"@type": "propertyValue"
},
{
"name": "birth_order",
"@type": "propertyValue"
},
{
"name": "count_birth_order",
"value": "1. 1/2,\n2. 2/2,\n3. 1/3,\n4. 2/3,\n5. 3/3,\n6. 1/4,\n7. 2/4,\n8. 3/4,\n9. 4/4,\n10. 1/5,\n11. 2/5,\n12. 3/5,\n13. 4/5,\n14. 5/5,\n15. 1/>5,\n16. 2/>5,\n17. 3/>5,\n18. 4/>5,\n19. 5/>5,\n20. >5/>5",
"@type": "propertyValue"
}
]
}`