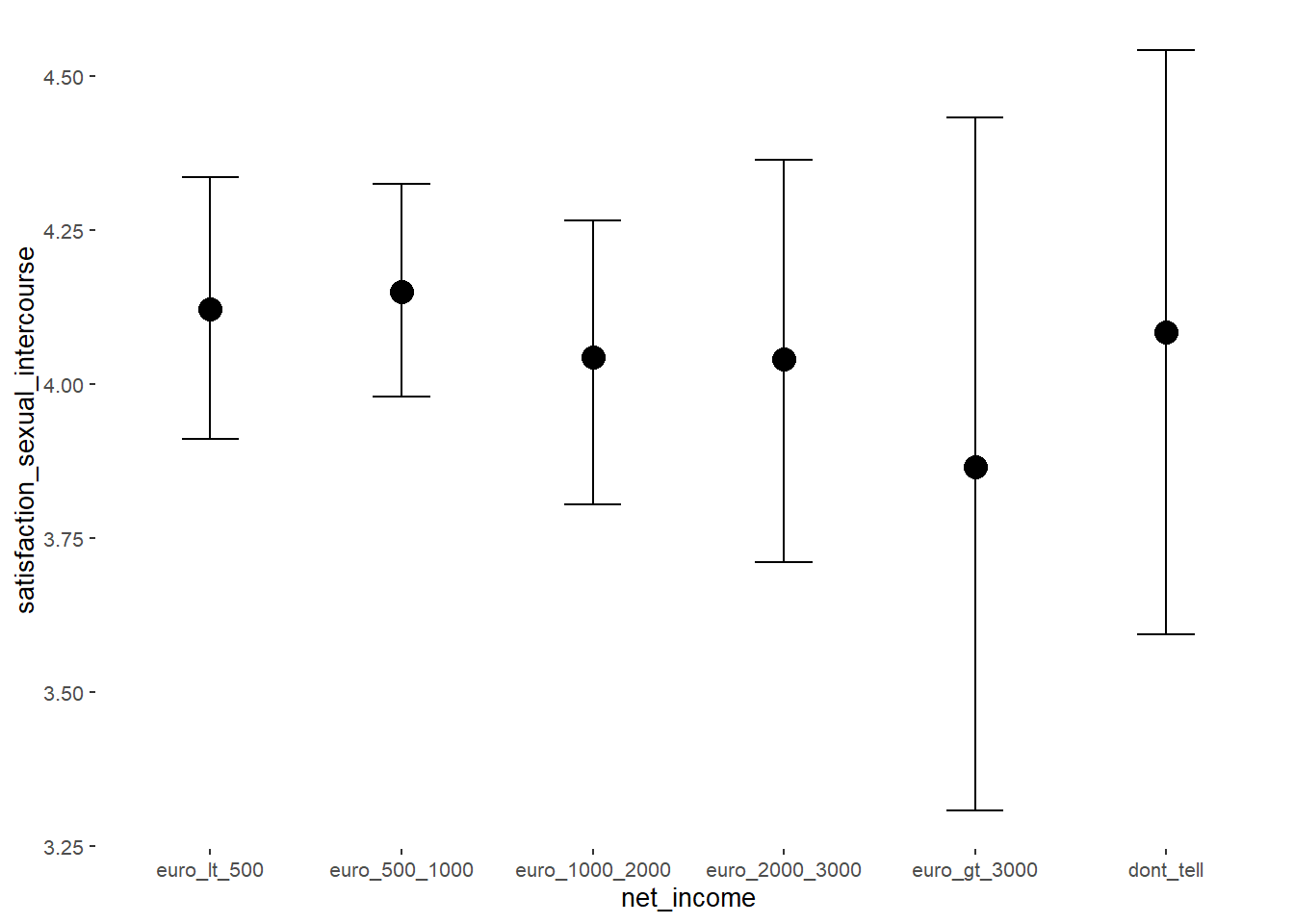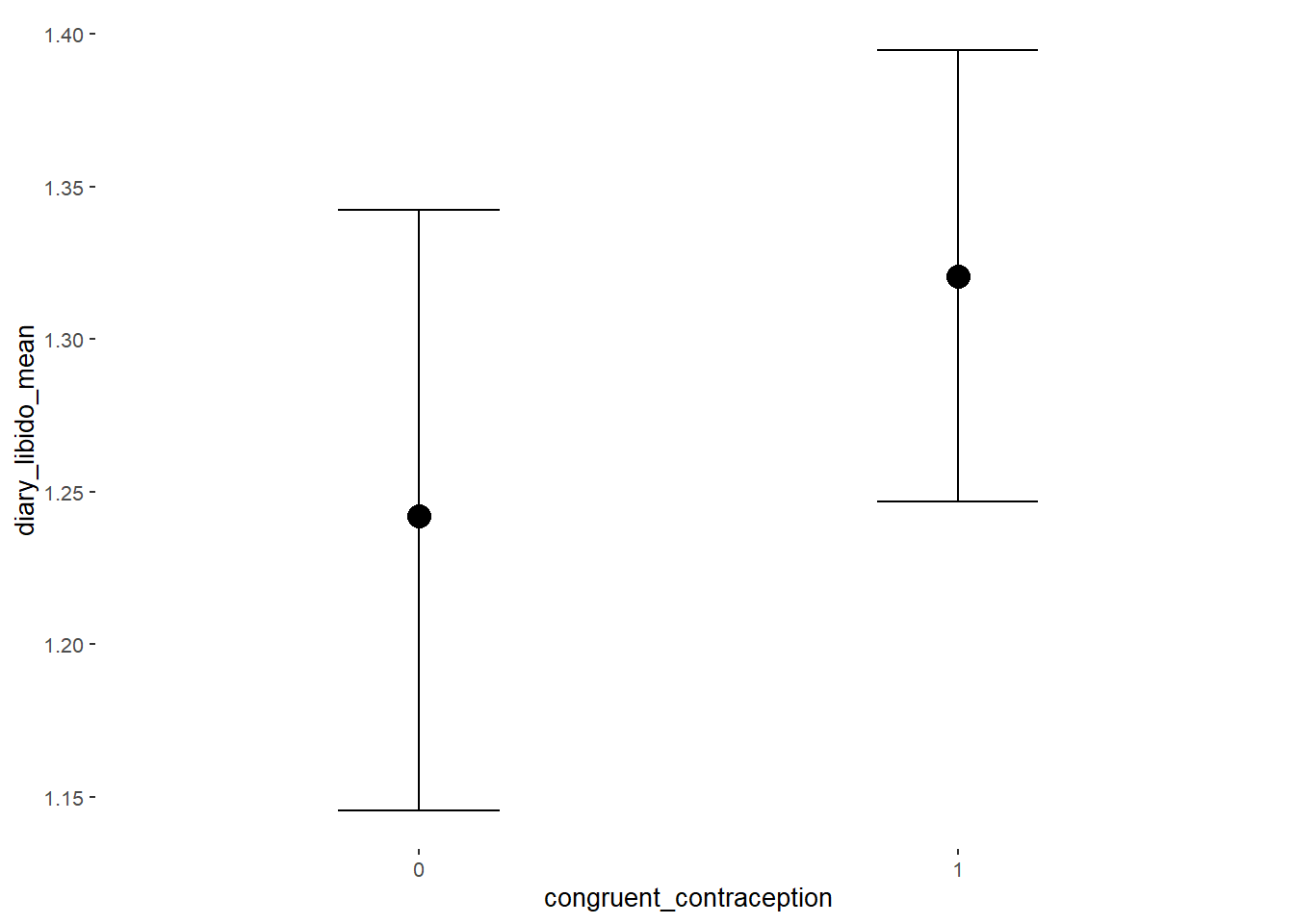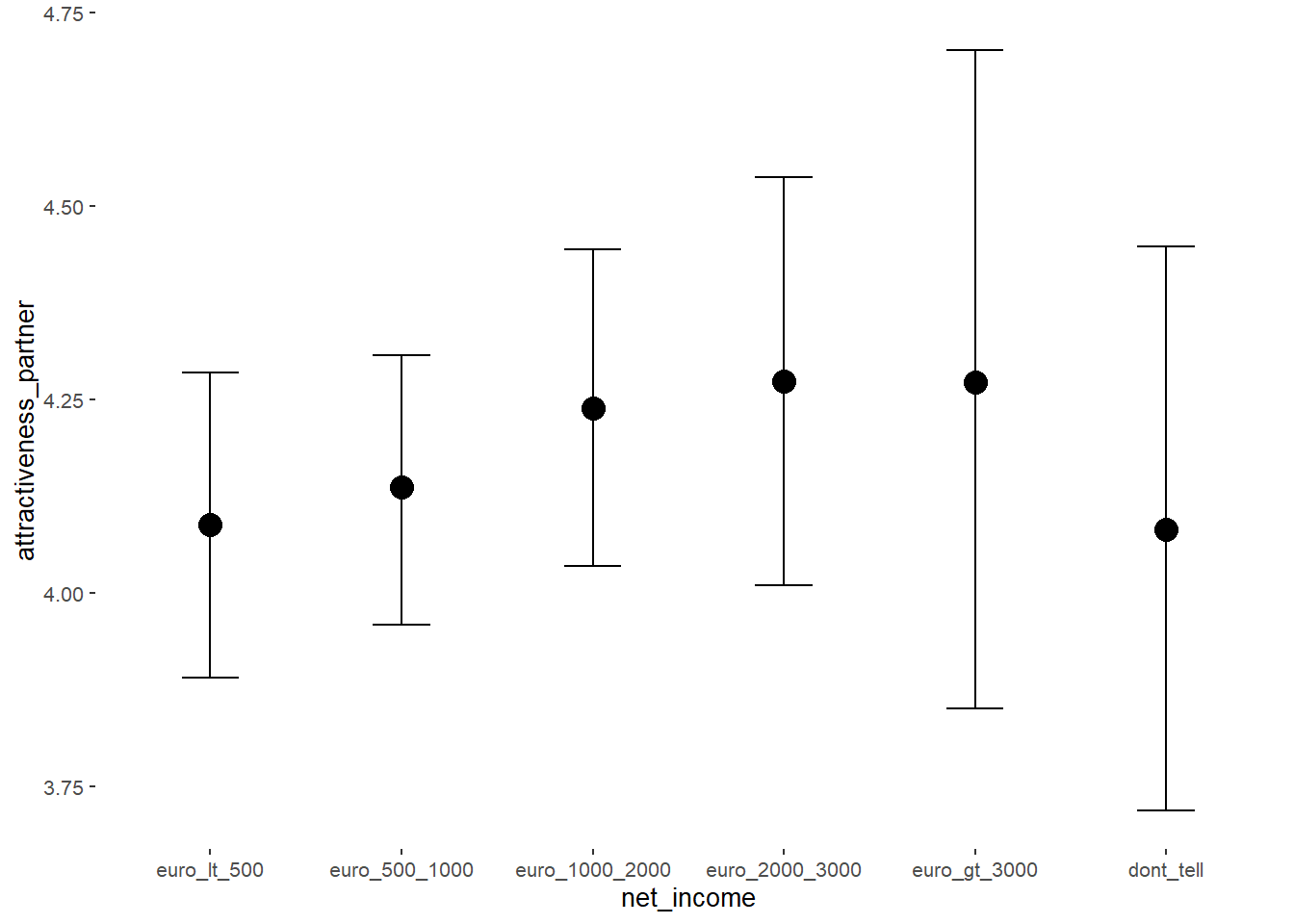Analyses Effects of Contraception
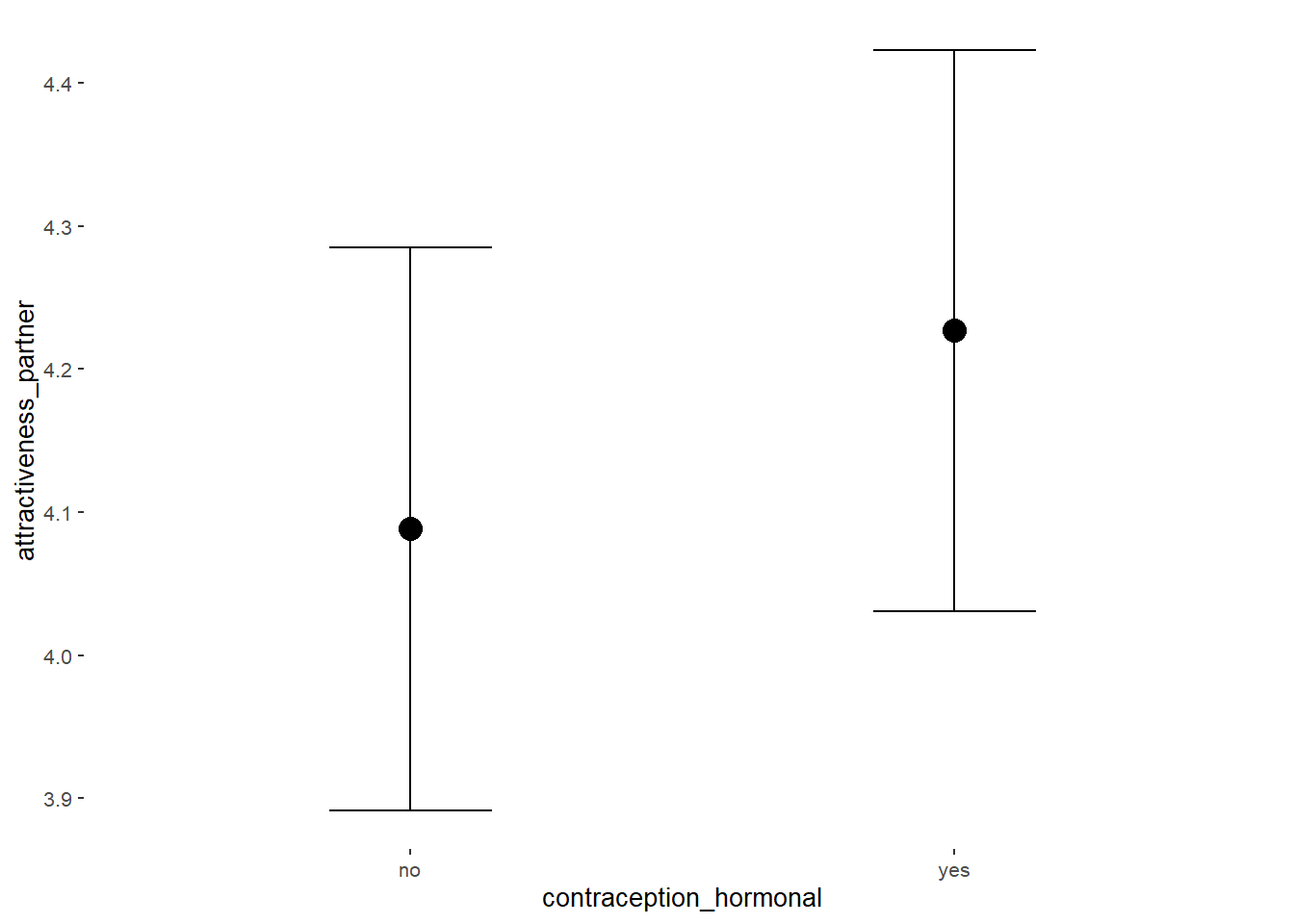
Effects of Hormonal Contraceptives
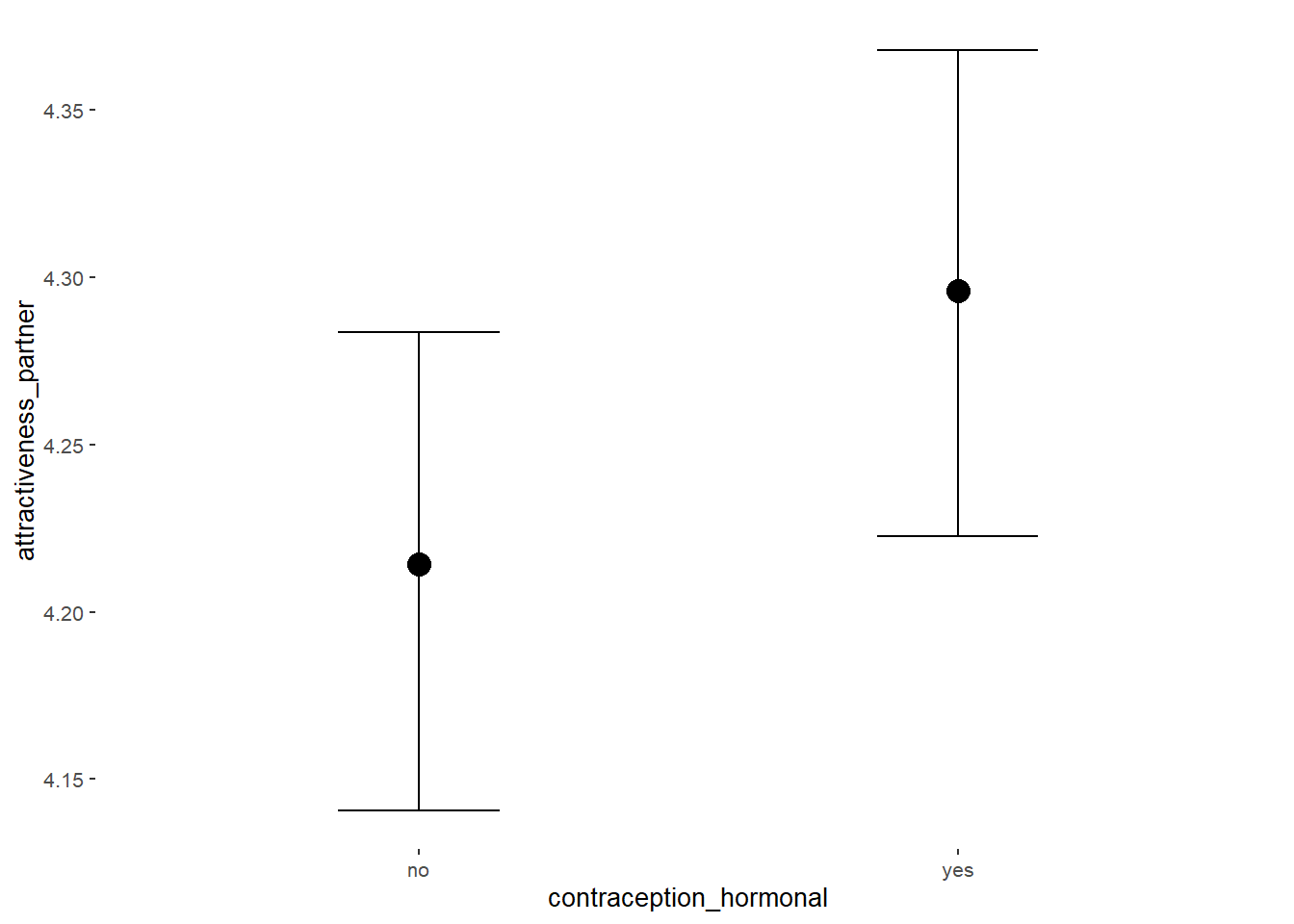
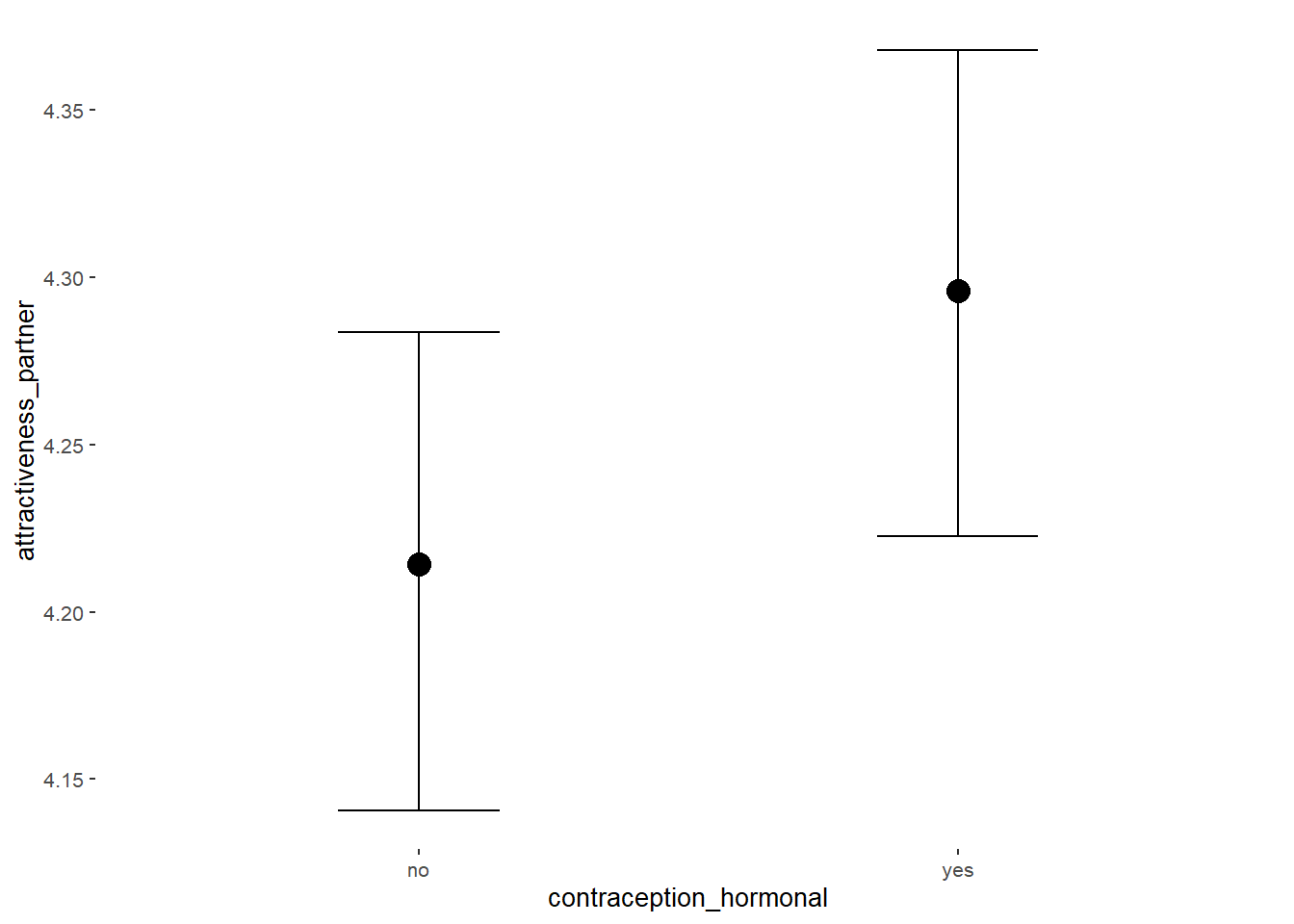
Attractiveness of Partner
Model
Summary
## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: attractiveness_partner ~ contraception_hormonal
## Data: data (Number of observations: 774)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## Intercept 4.21 0.04 4.15 4.27 1.00 3926 2990
## contraception_hormonalyes 0.08 0.05 -0.00 0.17 1.00 3780 2987
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.74 0.02 0.71 0.77 1.00 4114 2621
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
## Picking joint bandwidth of 0.00731## Warning: Removed 399 rows containing non-finite values (stat_density_ridges).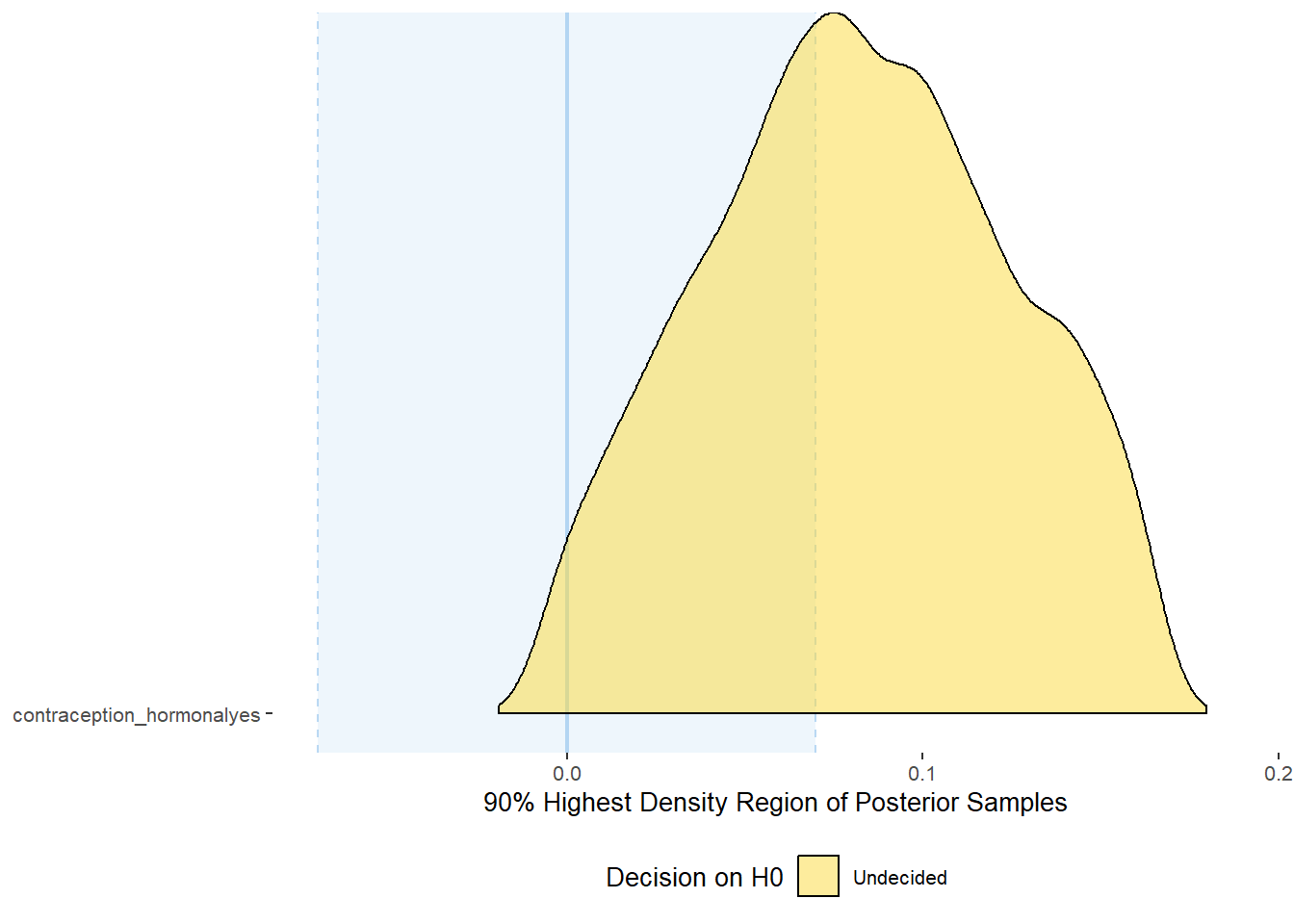
## # A tibble: 1 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contracepti~ 0.9 -0.07 0.07 0.400 Undecided -0.00626 0.166 fixed conditio~Forest Plot for Effect Sizes
m_hc_atrr %>%
spread_draws(b_contraception_hormonalyes) %>%
pivot_longer(cols = c(b_contraception_hormonalyes),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_contraception_hormonalyes",
"Contraception", NA),
condition = ifelse(condition %contains% "b_contraception_hormonalyes",
"Hormonal Contraception", NA)) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.07))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.07, 0.07), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors") +
xlim (-0.6, 0.6)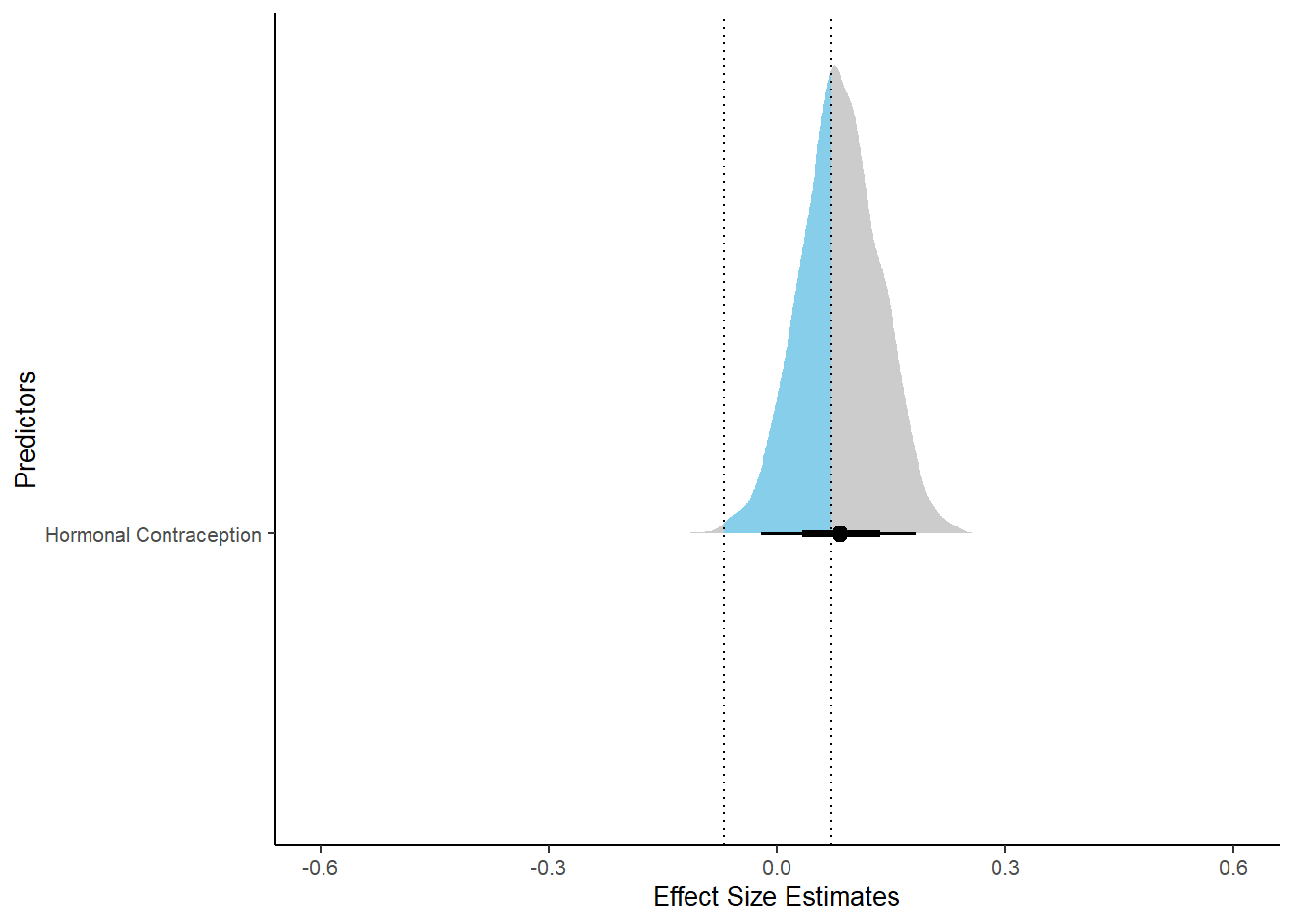
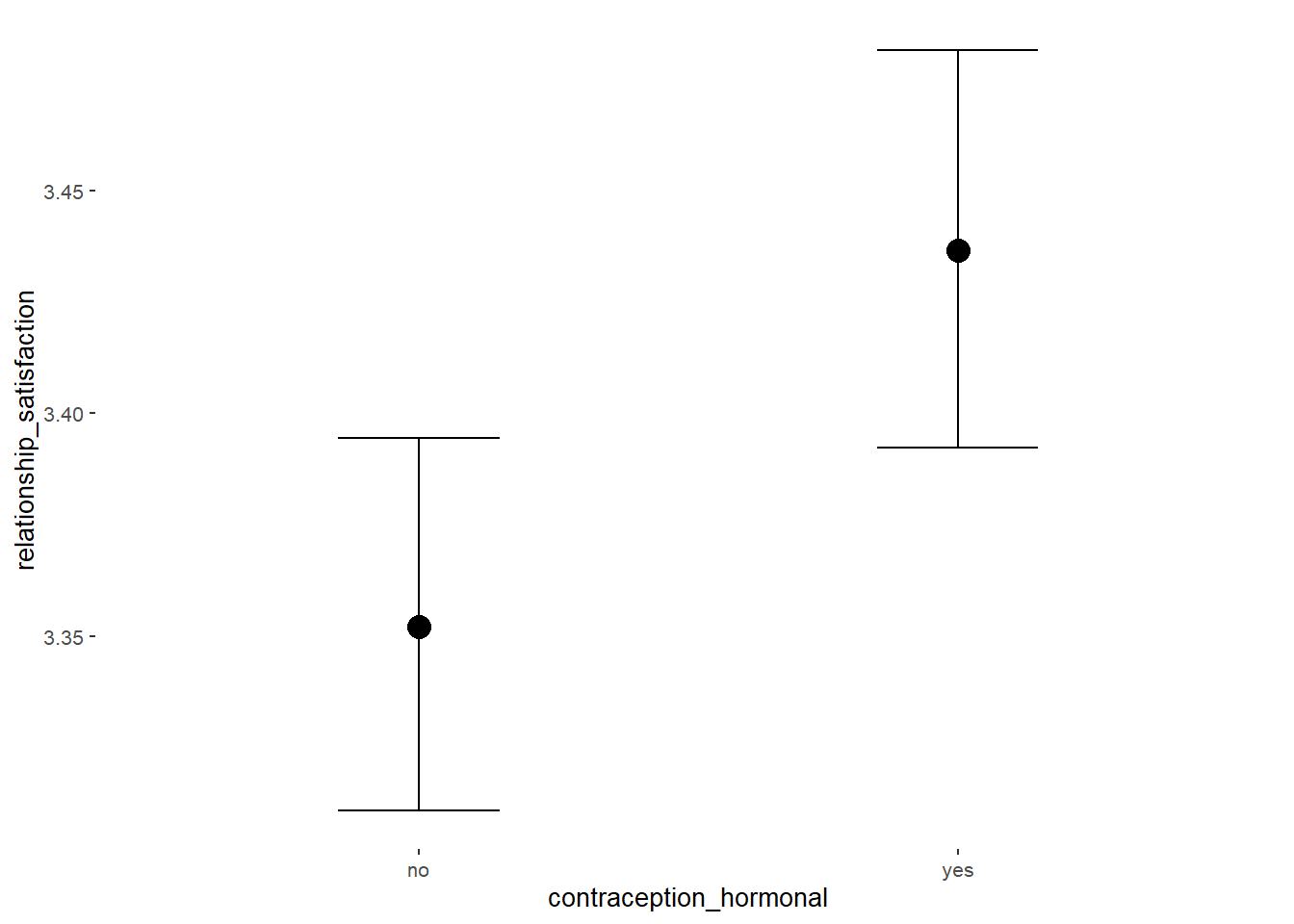
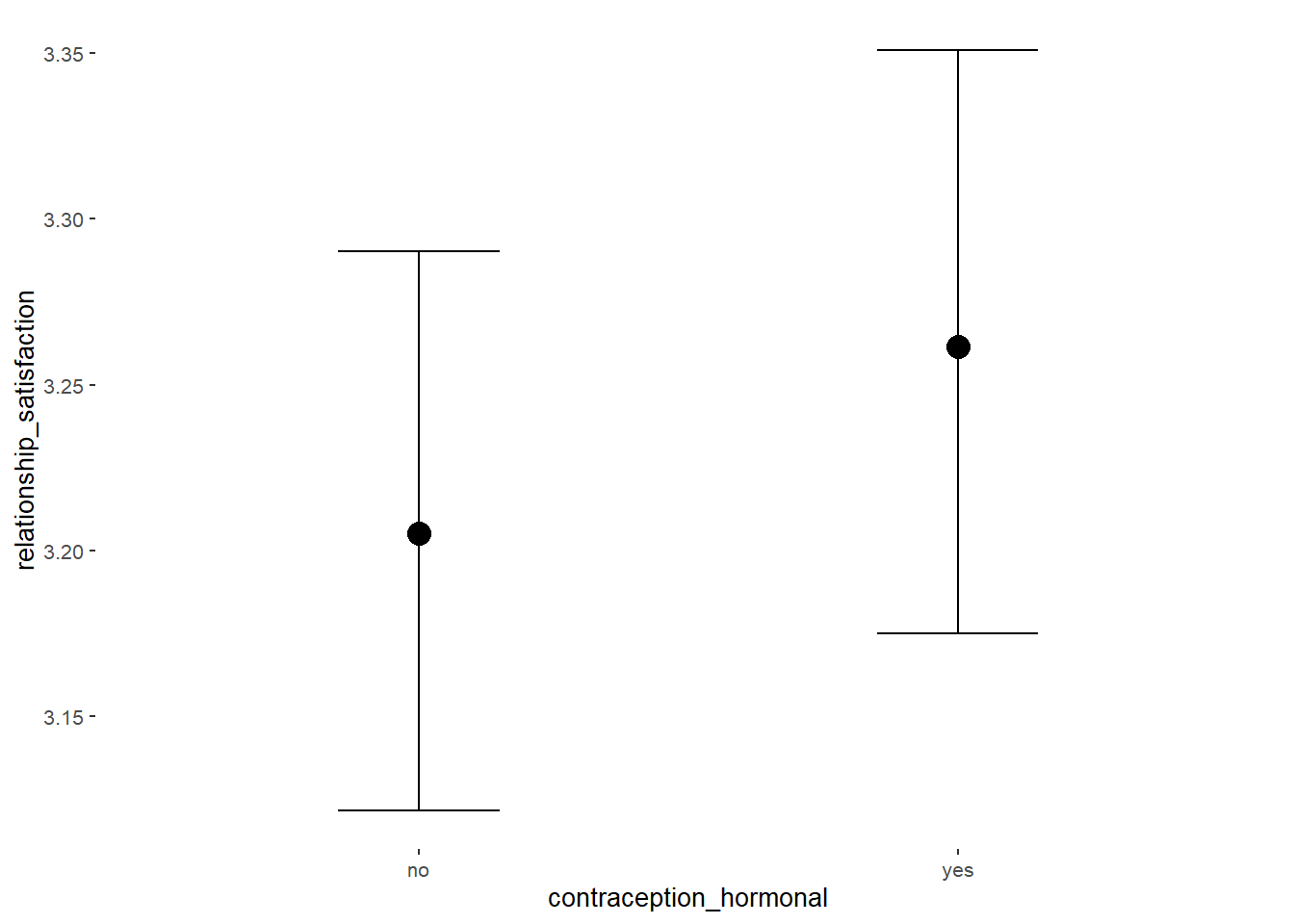
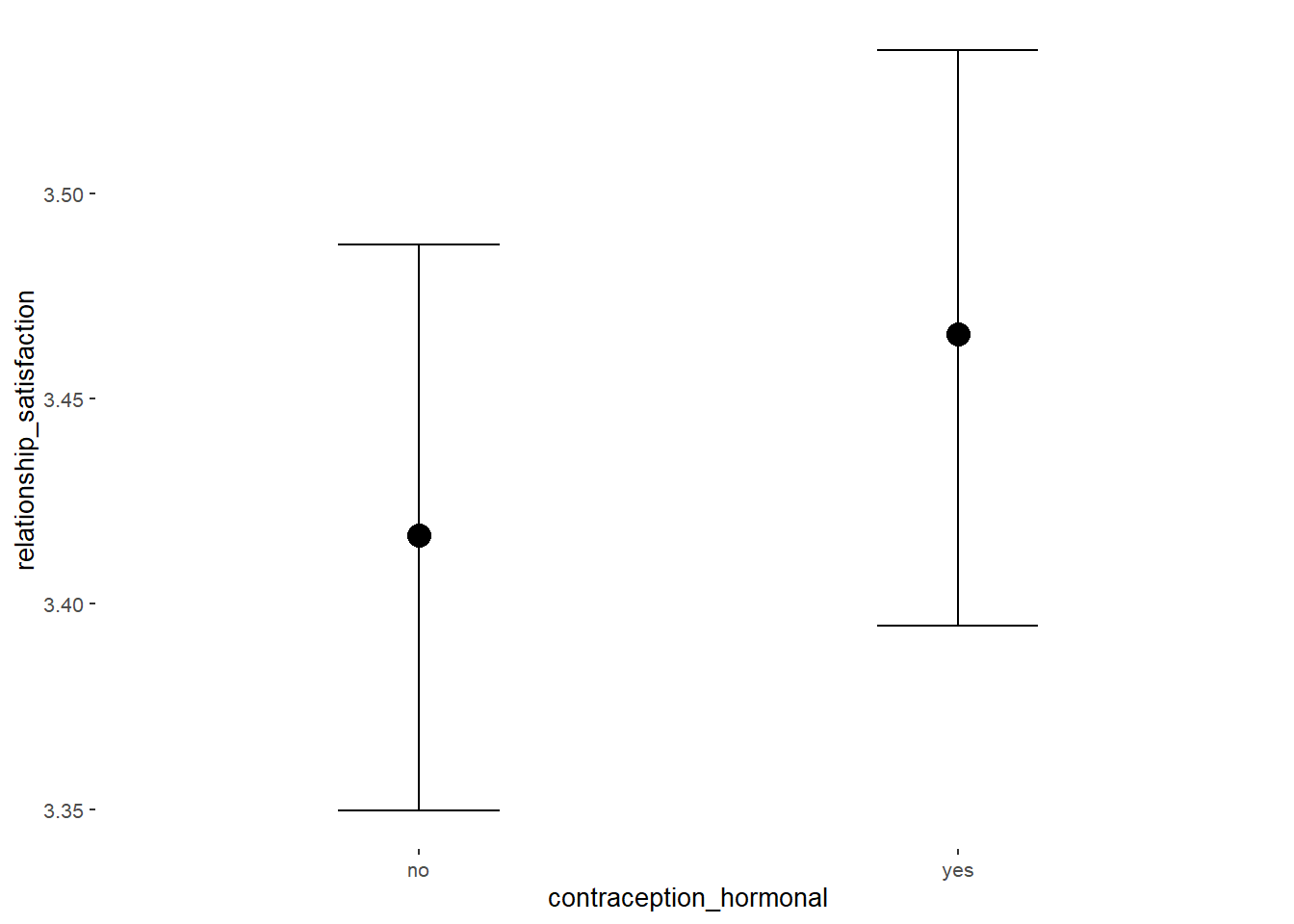
Relationship Satisfaction
Model
Summary
## Family: gaussian
## Links: mu = identity; sigma = identity
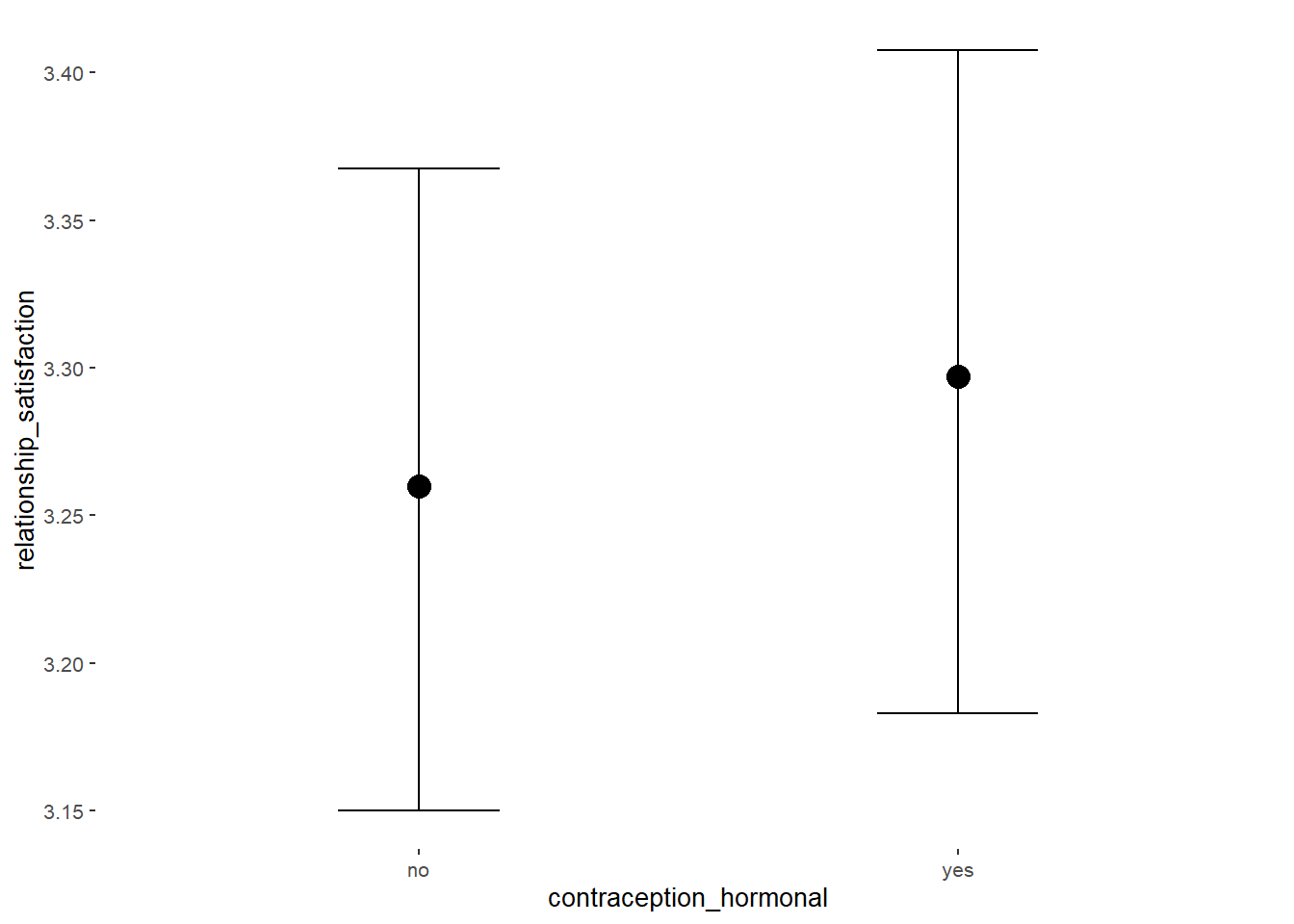
## Formula: relationship_satisfaction ~ contraception_hormonal
## Data: data (Number of observations: 774)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## Intercept 3.35 0.02 3.32 3.39 1.00 4070 2976
## contraception_hormonalyes 0.08 0.03 0.03 0.14 1.00 4119 2869
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.42 0.01 0.41 0.44 1.00 4602 2782
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_hc_relsat, range = c(-0.04, 0.04), ci = 0.90,
parameters = "contraception"))## Picking joint bandwidth of 0.00438## Warning: Removed 399 rows containing non-finite values (stat_density_ridges).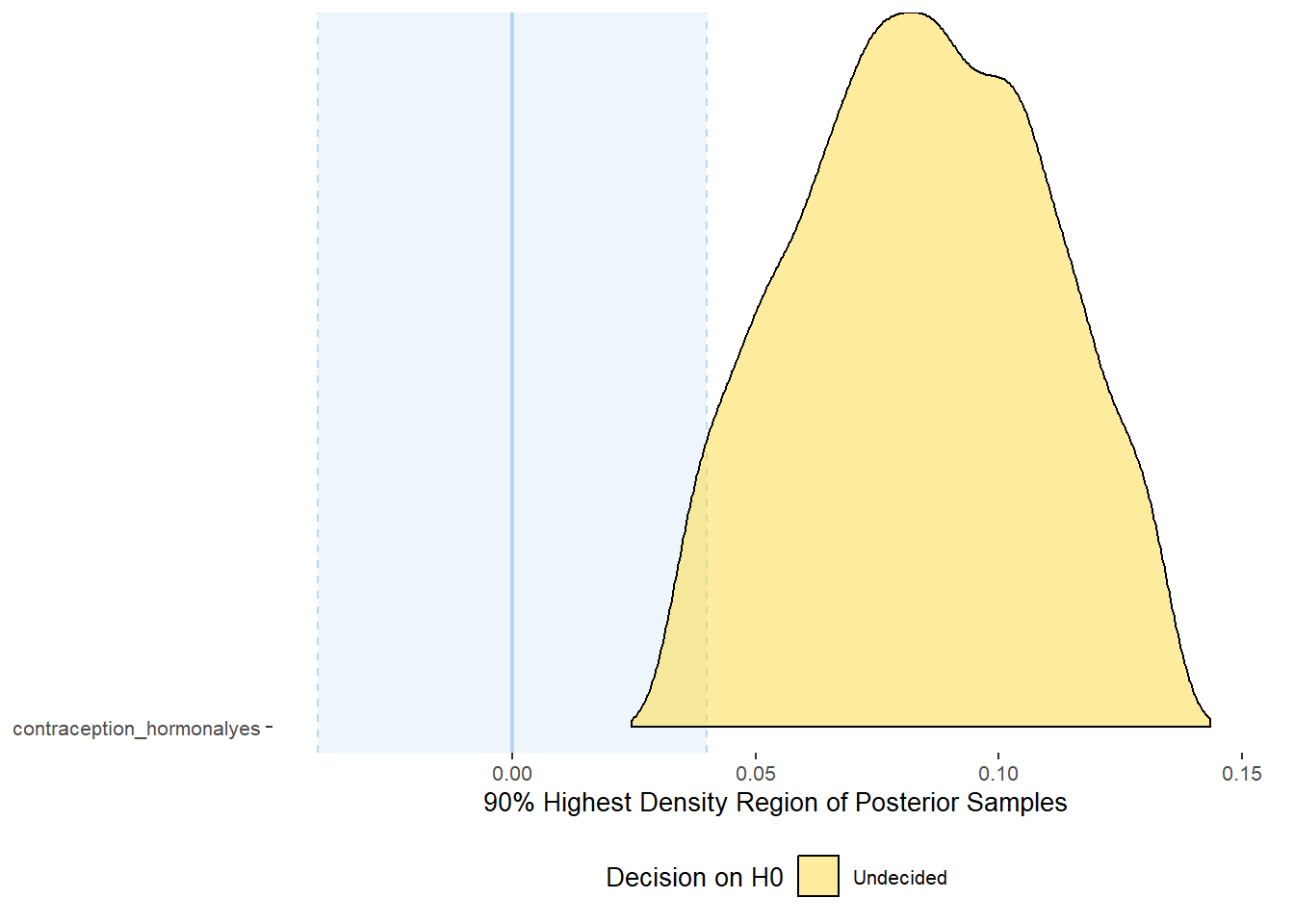
## # A tibble: 1 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.04 0.04 0.0361 Undecided 0.0325 0.135 fixed conditio~Forest Plot for Effect Sizes
m_hc_relsat %>%
spread_draws(b_contraception_hormonalyes) %>%
pivot_longer(cols = c(b_contraception_hormonalyes),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_contraception_hormonalyes",
"Contraception", NA),
condition = ifelse(condition %contains% "b_contraception_hormonalyes",
"Hormonal Contraception", NA)) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.04))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.04, 0.04), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors") +
xlim (-0.6, 0.6)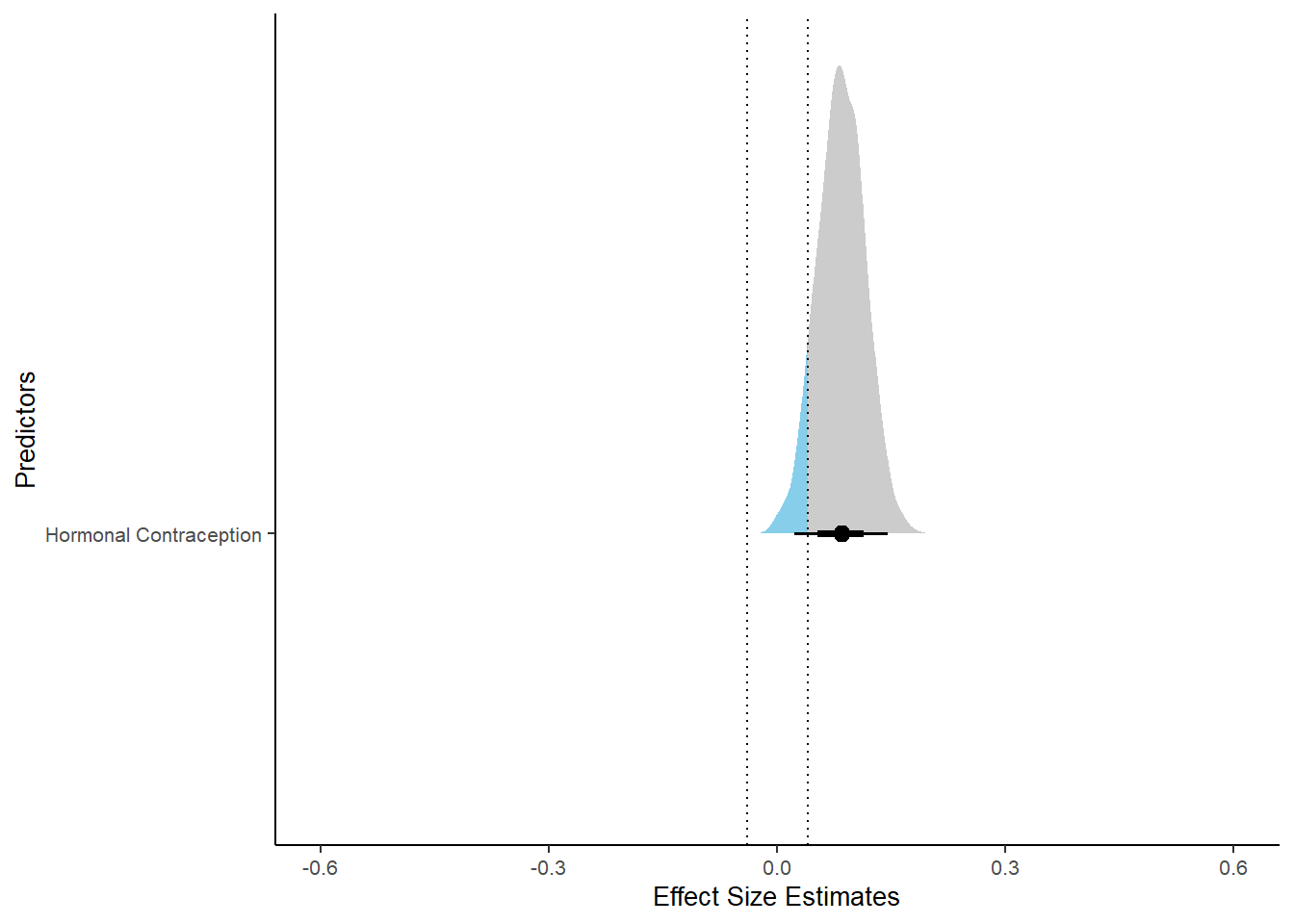
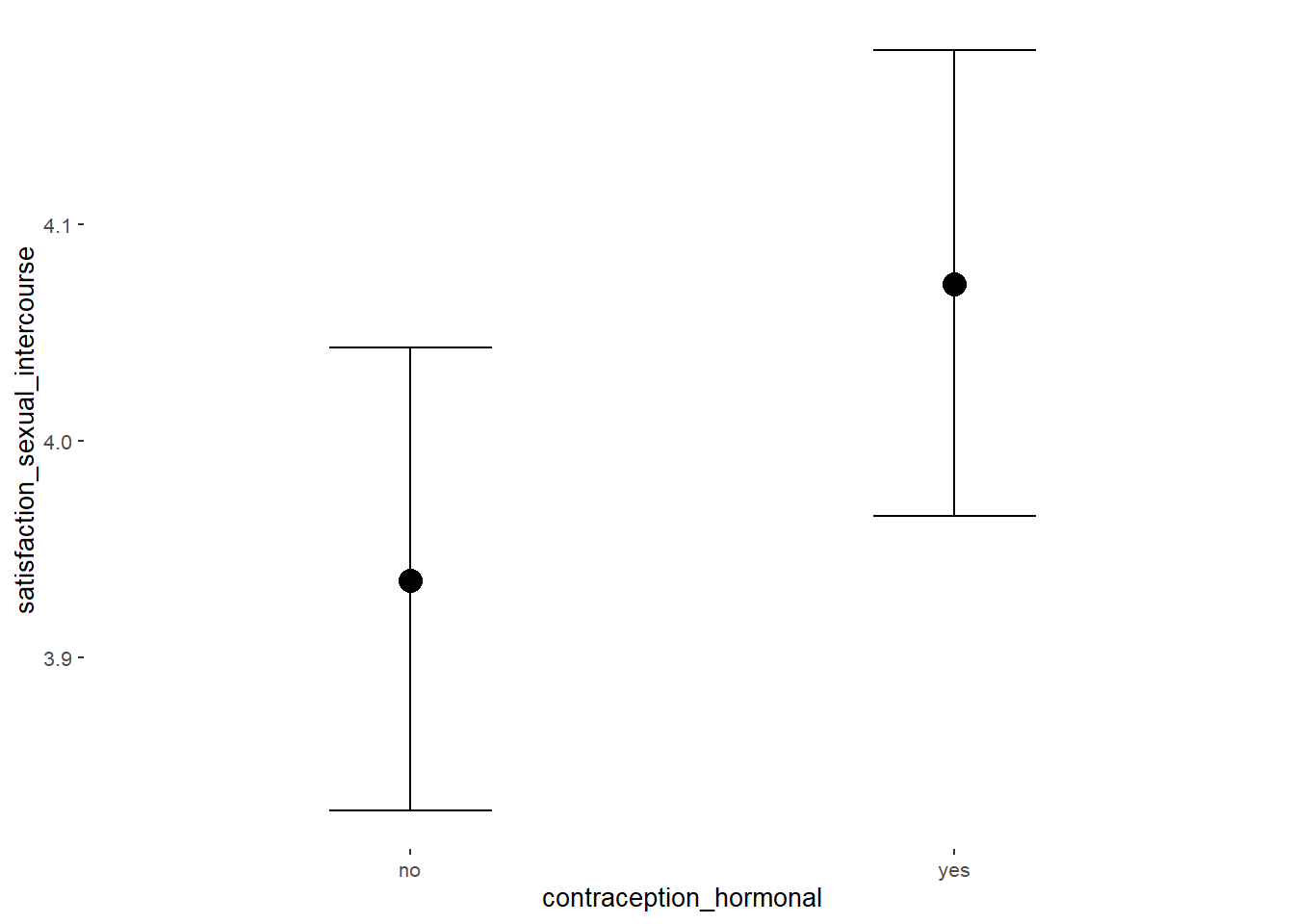
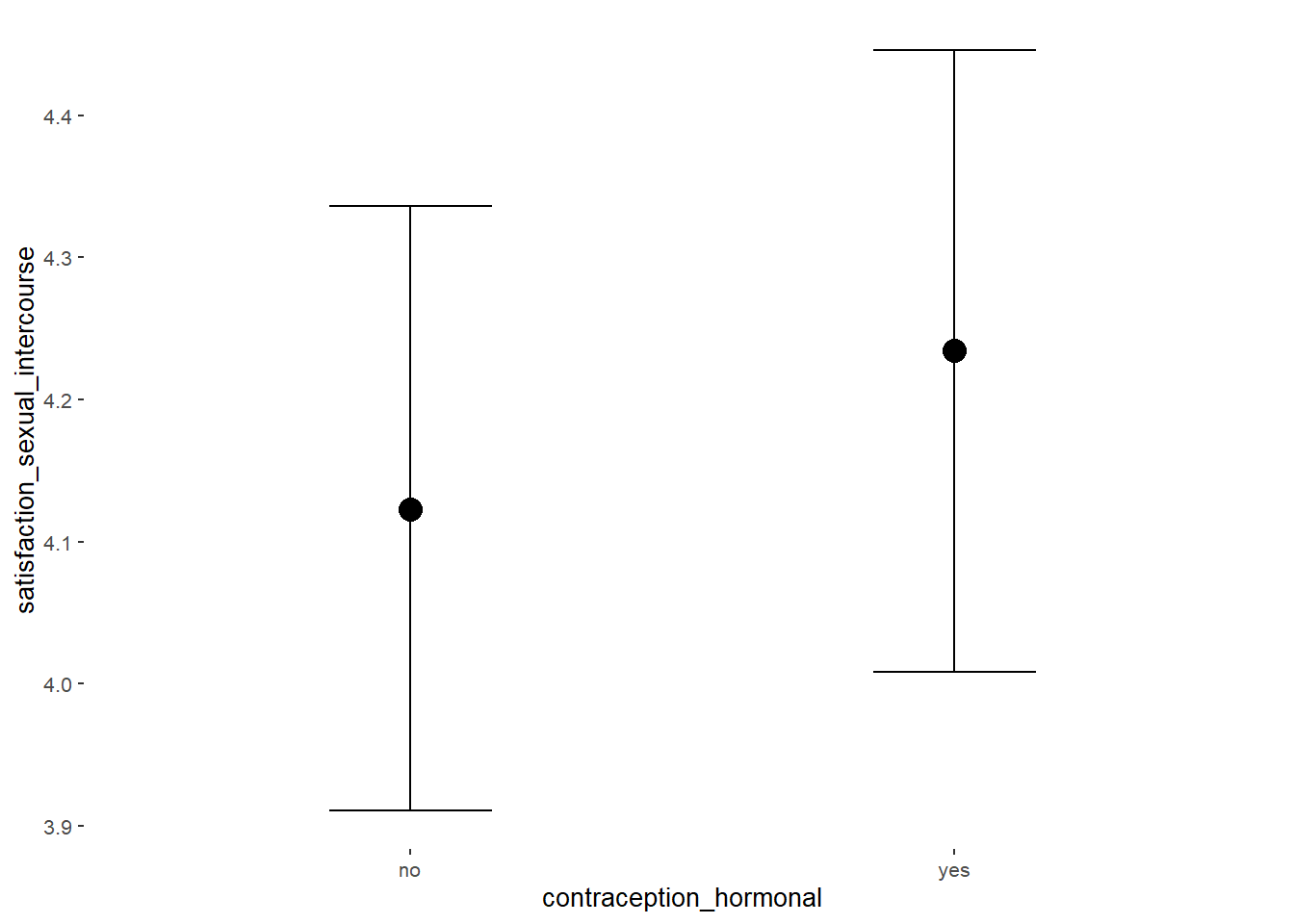
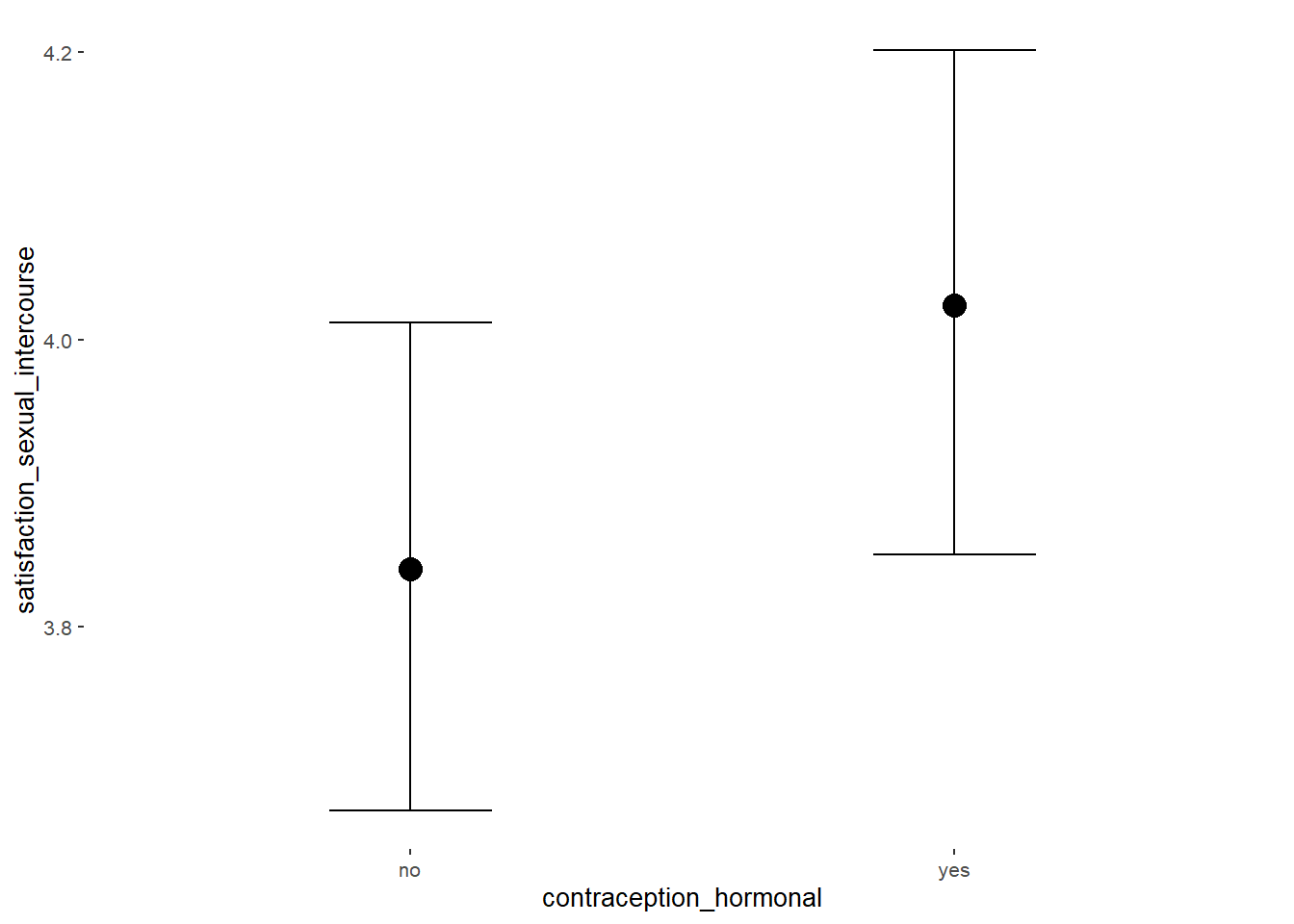
Sexual Satisfaction
Model
Summary
## Family: gaussian
## Links: mu = identity; sigma = identity
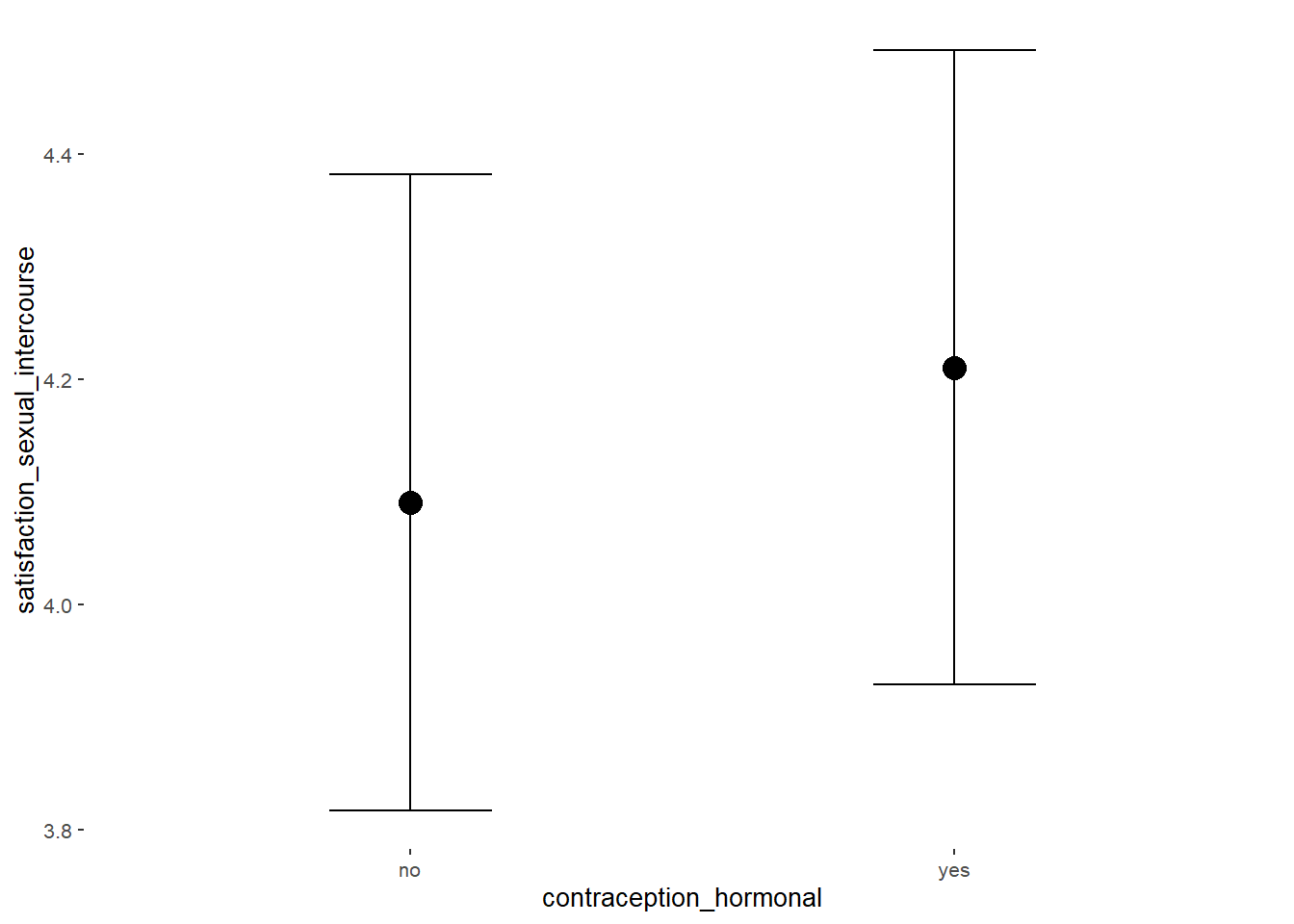
## Formula: satisfaction_sexual_intercourse ~ contraception_hormonal
## Data: data (Number of observations: 774)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## Intercept 3.94 0.05 3.84 4.03 1.00 4000 2867
## contraception_hormonalyes 0.14 0.08 0.01 0.26 1.00 4133 3129
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 1.05 0.03 1.01 1.10 1.00 4281 2820
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
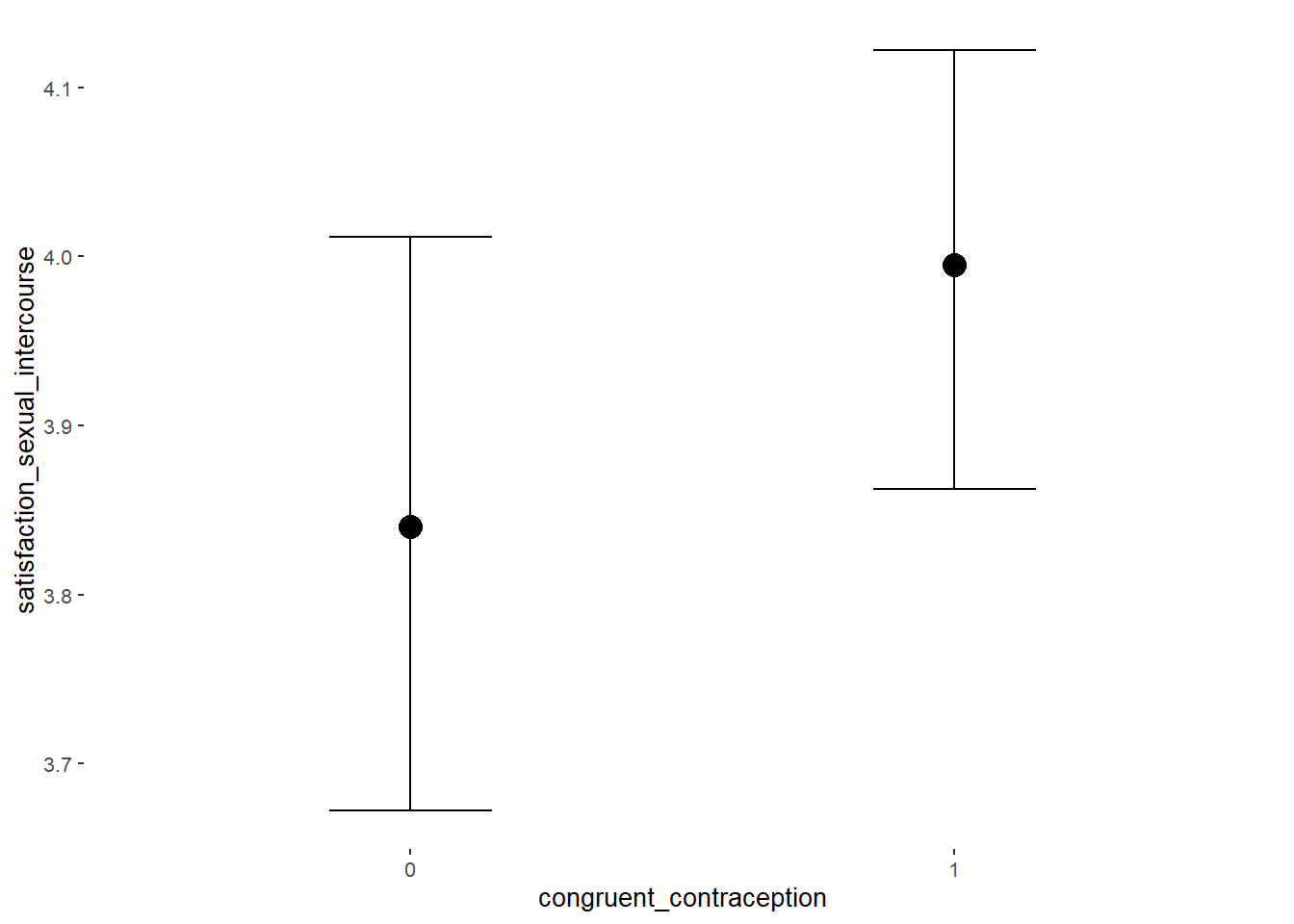
plot(equivalence_test(m_hc_sexsat, range = c(-0.11, 0.11), ci = 0.90,
parameters = "contraception"))## Picking joint bandwidth of 0.0106## Warning: Removed 399 rows containing non-finite values (stat_density_ridges).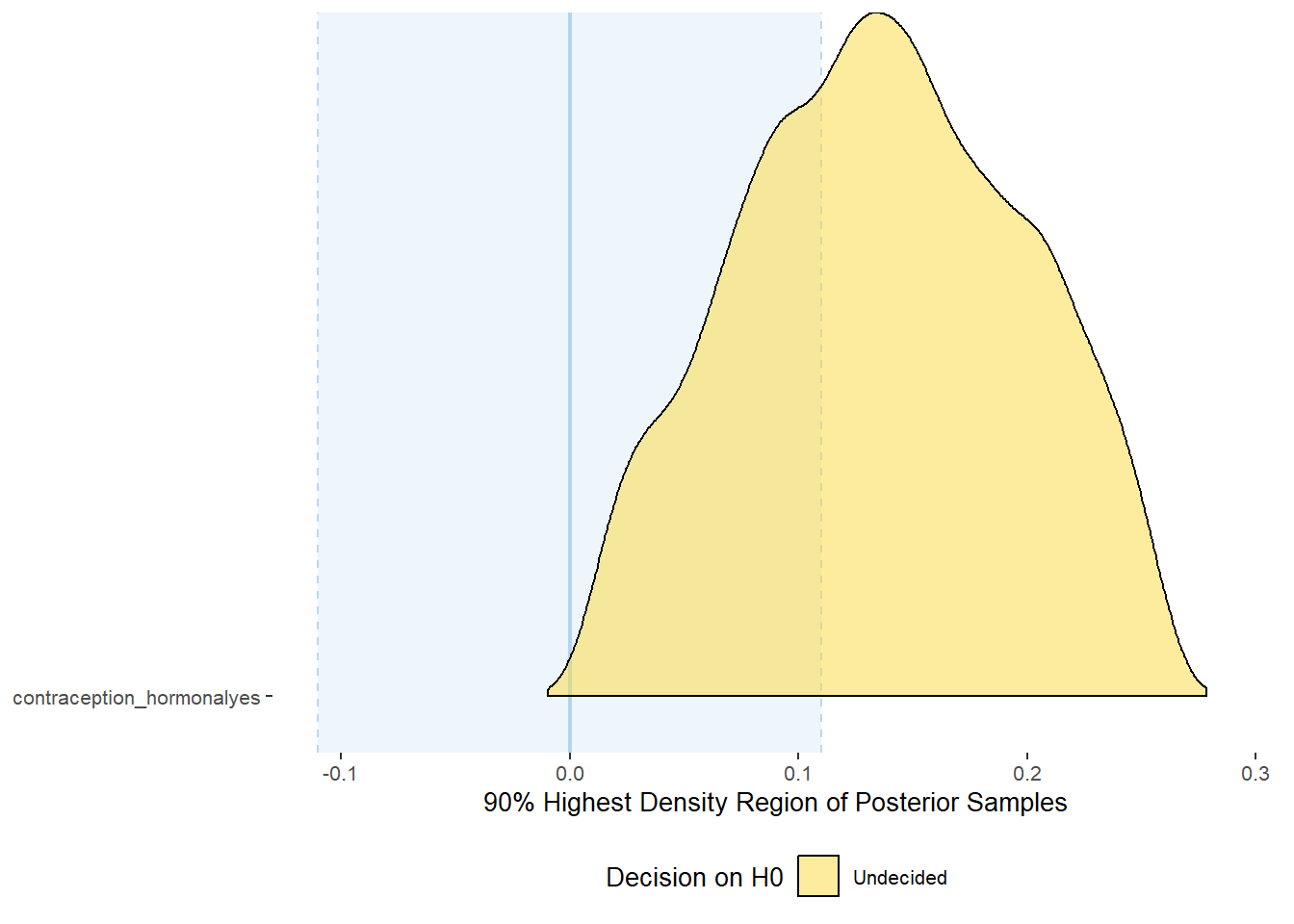
## # A tibble: 1 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.11 0.11 0.347 Undecided 0.00914 0.259 fixed conditio~Forest Plot for Effect Sizes
m_hc_sexsat %>%
spread_draws(b_contraception_hormonalyes) %>%
pivot_longer(cols = c(b_contraception_hormonalyes),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_contraception_hormonalyes",
"Contraception", NA),
condition = ifelse(condition %contains% "b_contraception_hormonalyes",
"Hormonal Contraception", NA)) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.11))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.11, 0.11), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors") +
xlim (-0.6, 0.6)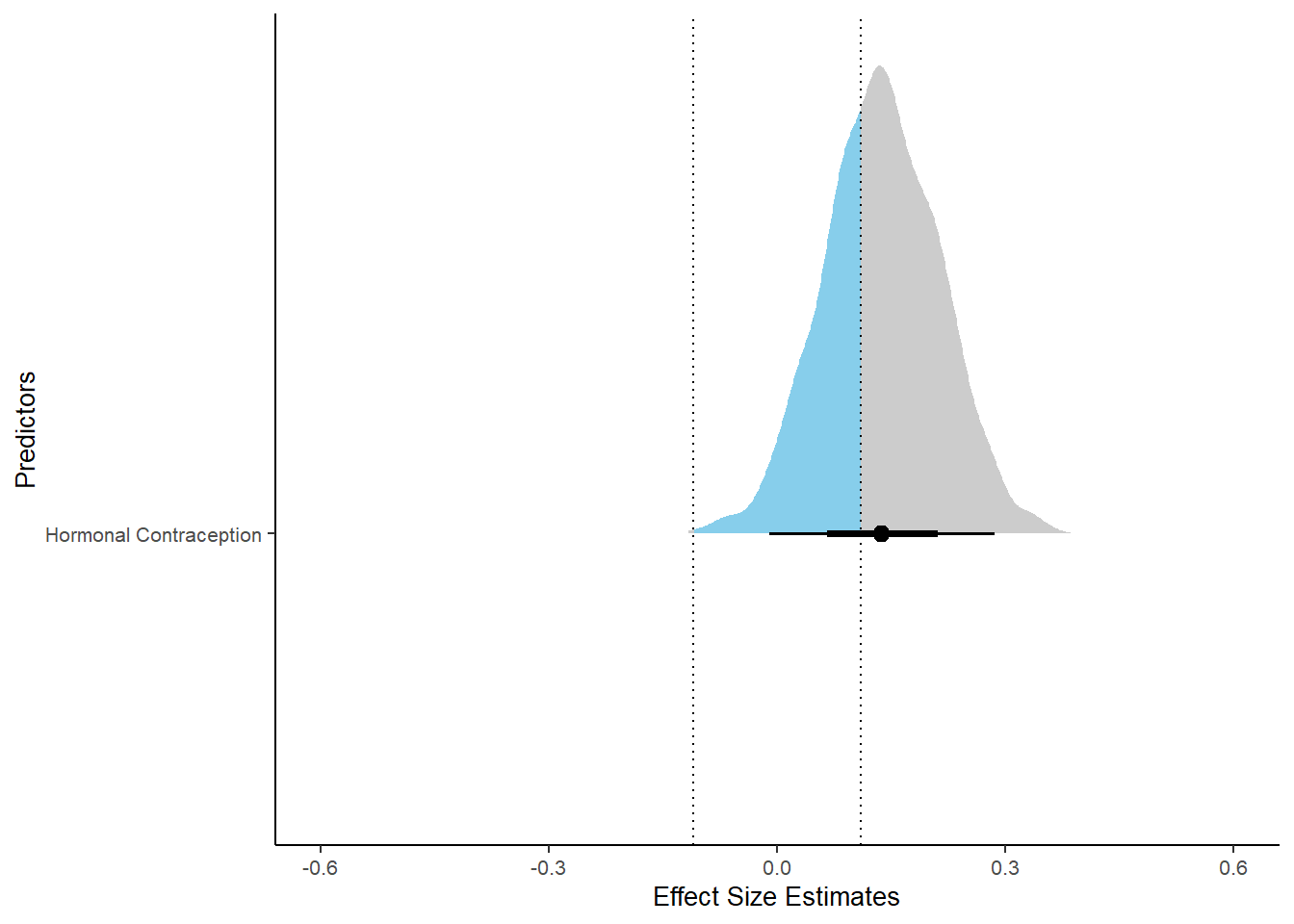
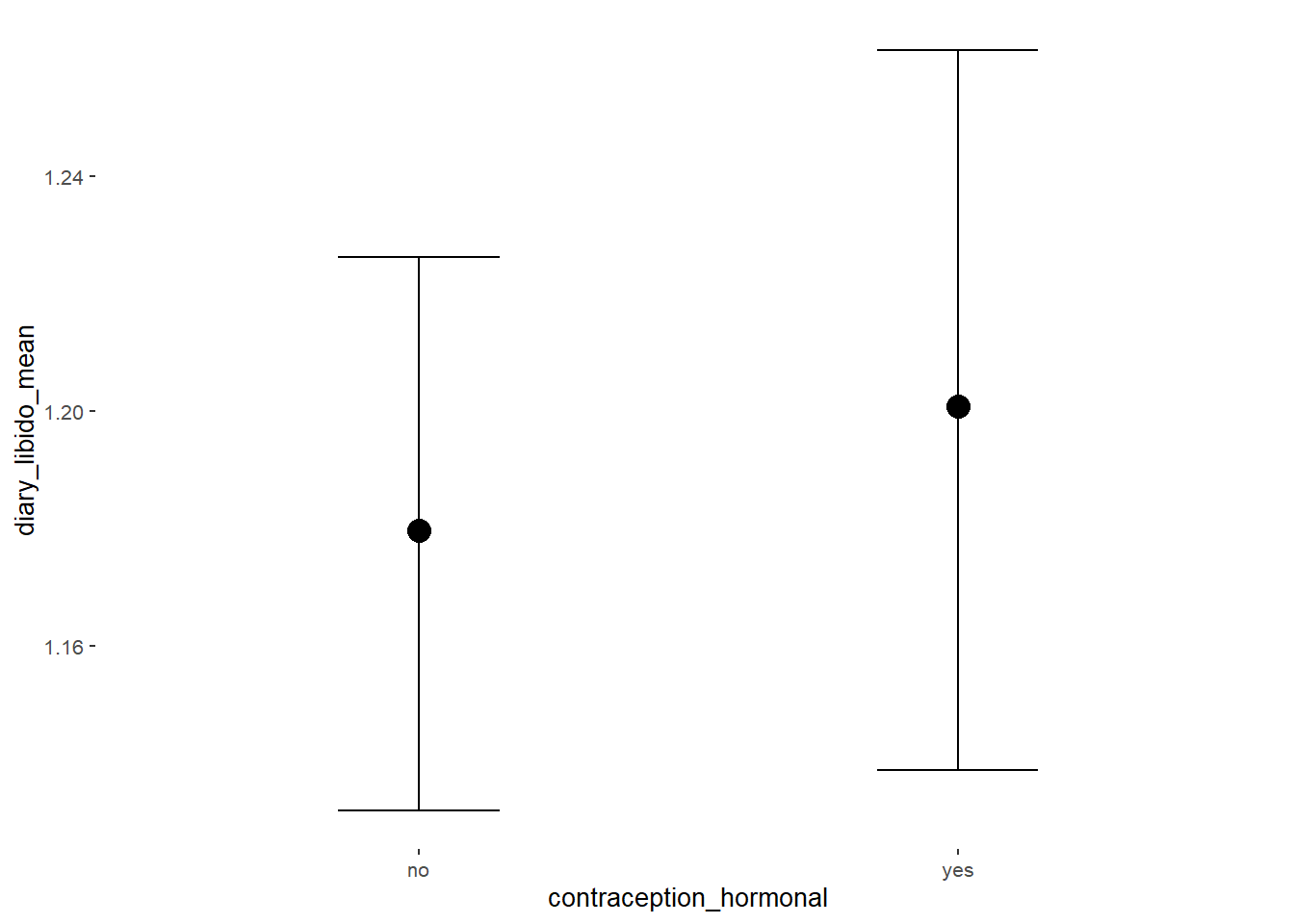
Libido
Model
Summary
## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: diary_libido_mean ~ contraception_hormonal
## Data: data (Number of observations: 968)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## Intercept 1.18 0.02 1.14 1.22 1.00 4104 3097
## contraception_hormonalyes 0.02 0.04 -0.04 0.08 1.00 4305 2788
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.59 0.01 0.57 0.61 1.00 3347 2731
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
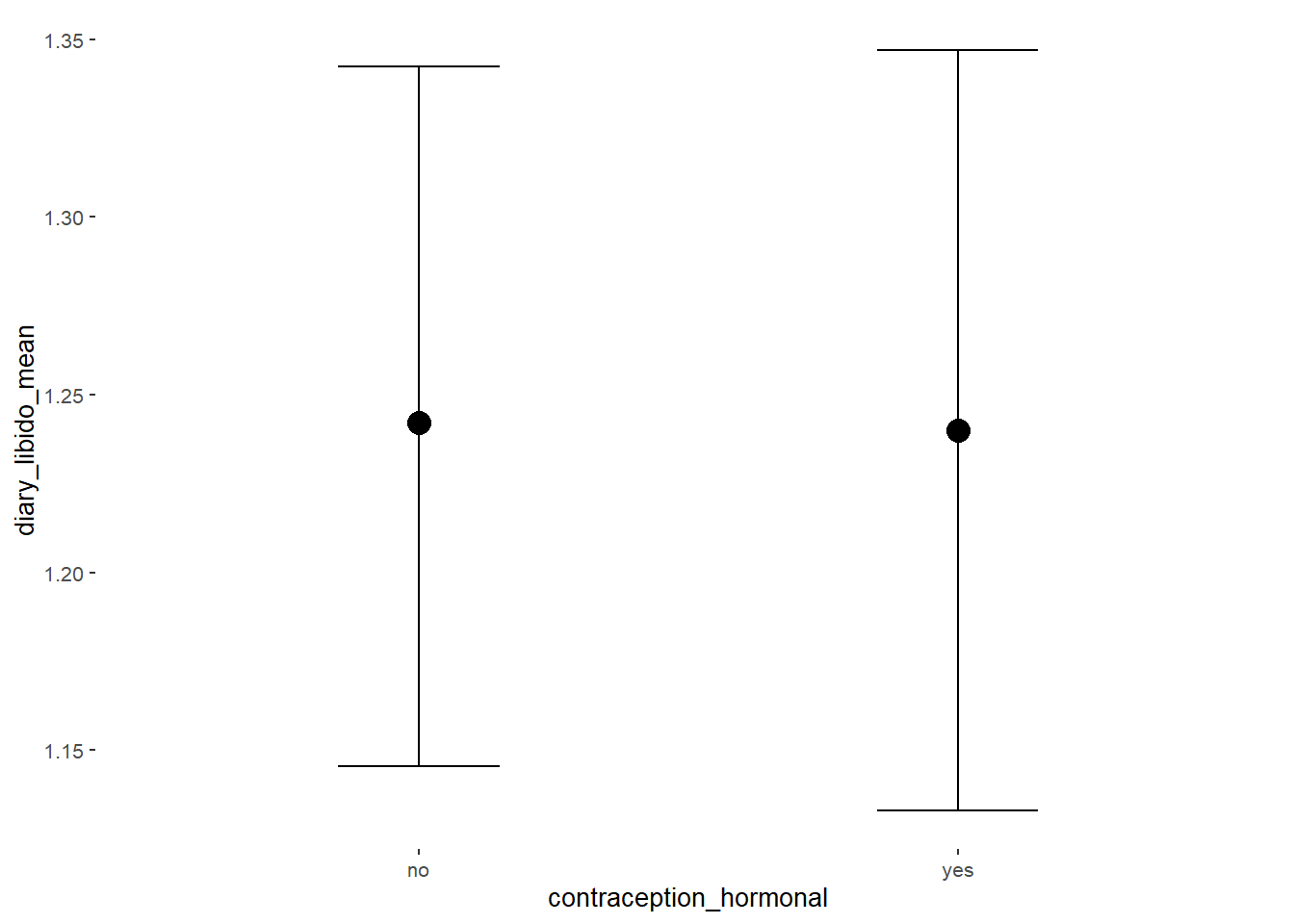
plot(equivalence_test(m_hc_libido, range = c(-0.06, 0.06), ci = 0.90,
parameters = "contraception"))## Picking joint bandwidth of 0.00544## Warning: Removed 399 rows containing non-finite values (stat_density_ridges).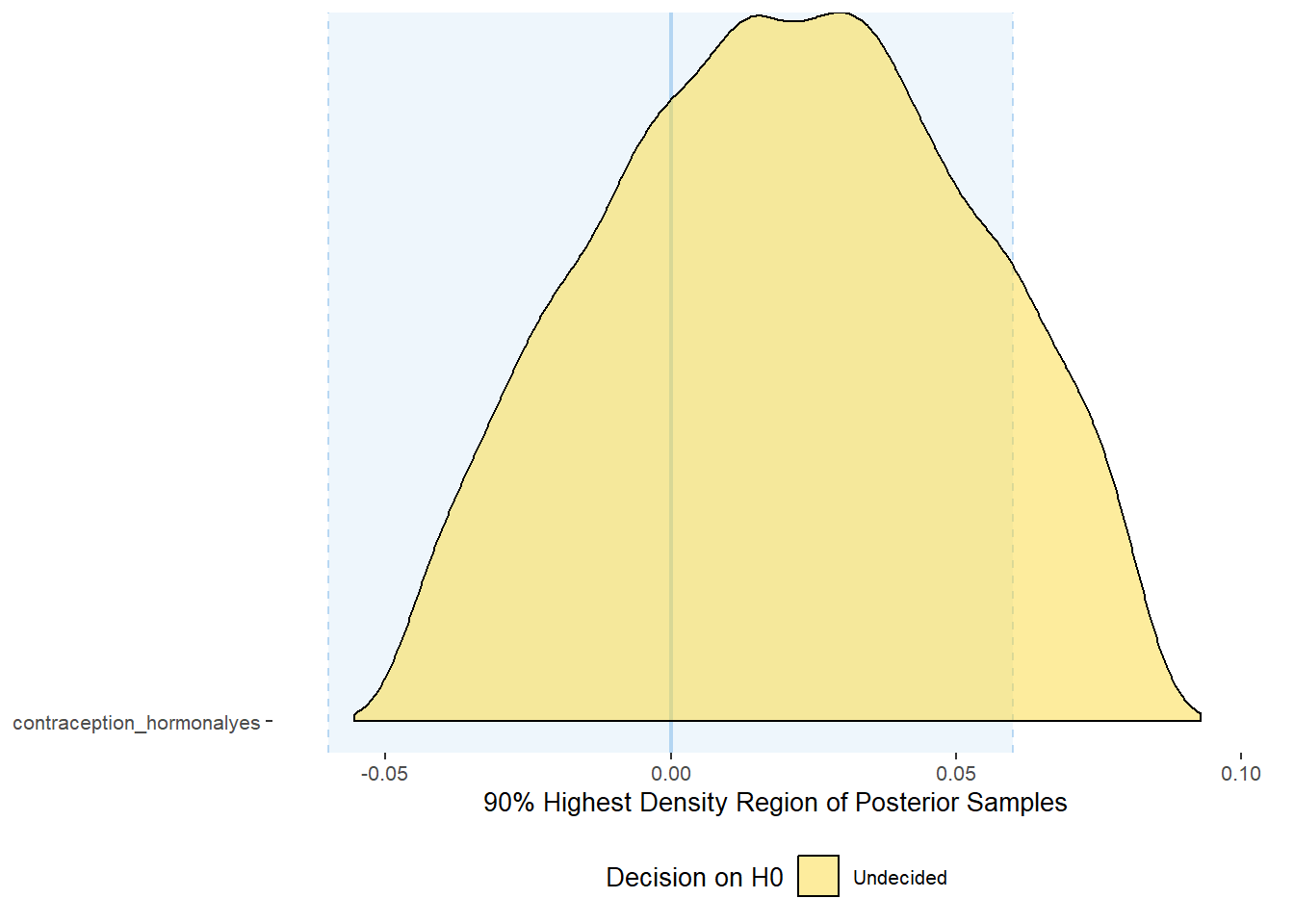
## # A tibble: 1 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.06 0.06 0.884 Undecided -0.0455 0.0826 fixed conditio~Forest Plot for Effect Sizes
m_hc_libido %>%
spread_draws(b_contraception_hormonalyes) %>%
pivot_longer(cols = c(b_contraception_hormonalyes),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_contraception_hormonalyes",
"Contraception", NA),
condition = ifelse(condition %contains% "b_contraception_hormonalyes",
"Hormonal Contraception", NA)) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.06))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.06, 0.06), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors") +
xlim (-0.6, 0.6)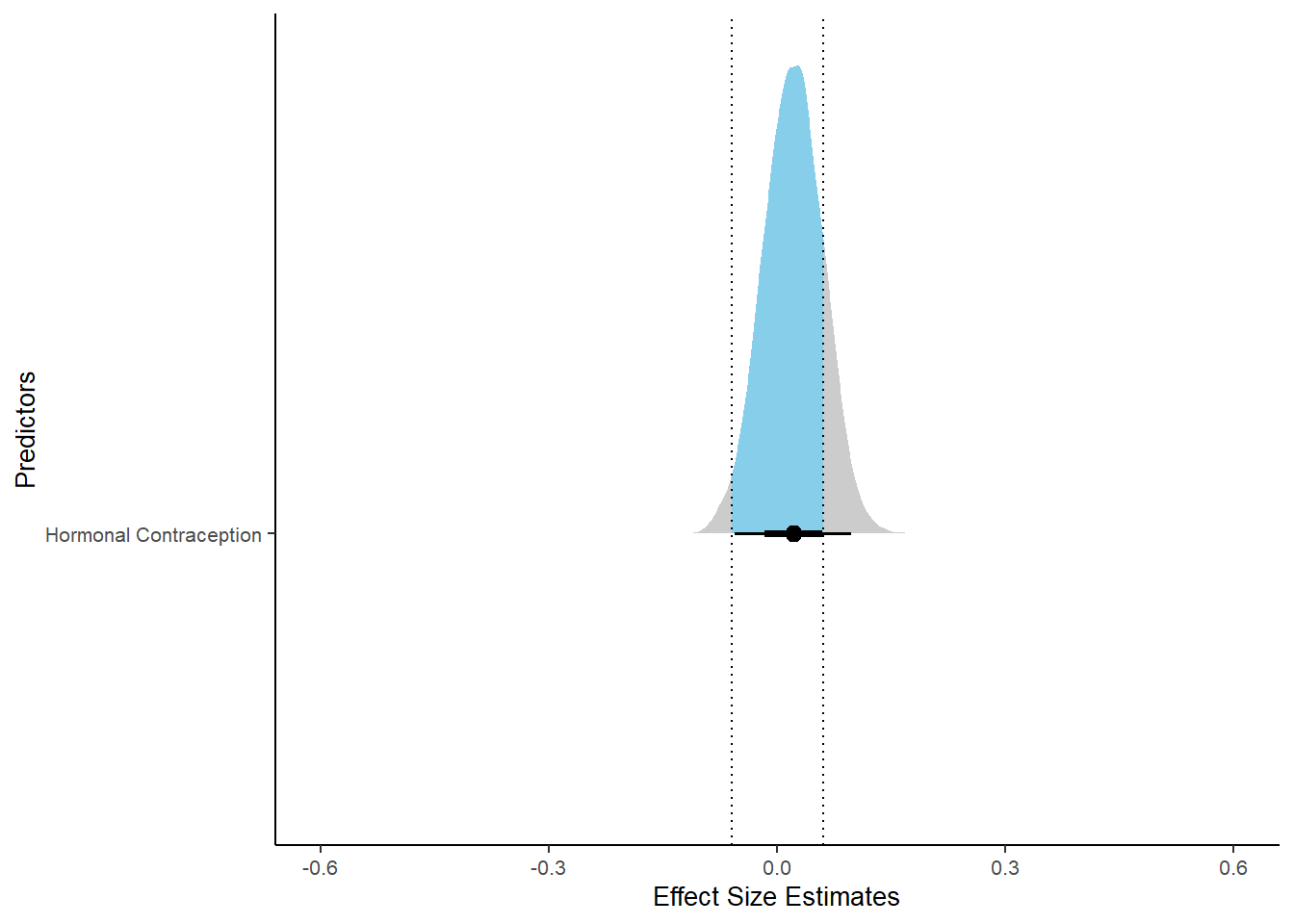
Sexual Frequency (Penetrative Intercourse)
Model
Summary
## Family: poisson
## Links: mu = log
## Formula: diary_sex_active_sex_sum ~ offset(log(number_of_days)) + contraception_hormonal
## Data: data (Number of observations: 897)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## Intercept -2.09 0.02 -2.12 -2.06 1.00 3349 2343
## contraception_hormonalyes 0.25 0.03 0.21 0.29 1.00 3880 2592
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_hc_sexfreqpen, range = c(-0.05, 0.05)), ci = 0.90,
parameters = "contraception")## Picking joint bandwidth of 0.00387## Warning: Removed 199 rows containing non-finite values (stat_density_ridges).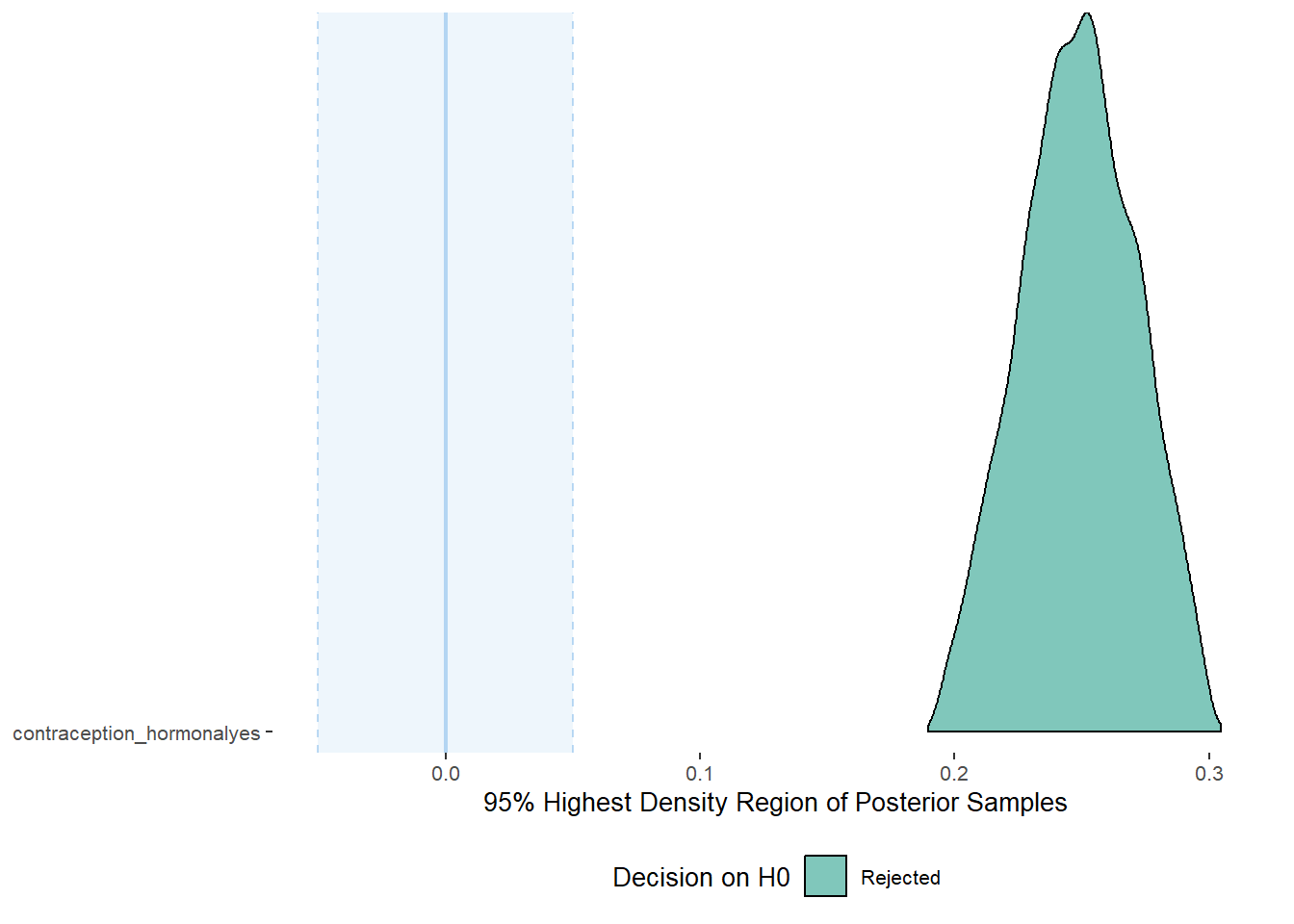
## # A tibble: 1 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.05 0.05 0 Rejected 0.205 0.290 fixed conditio~Plots
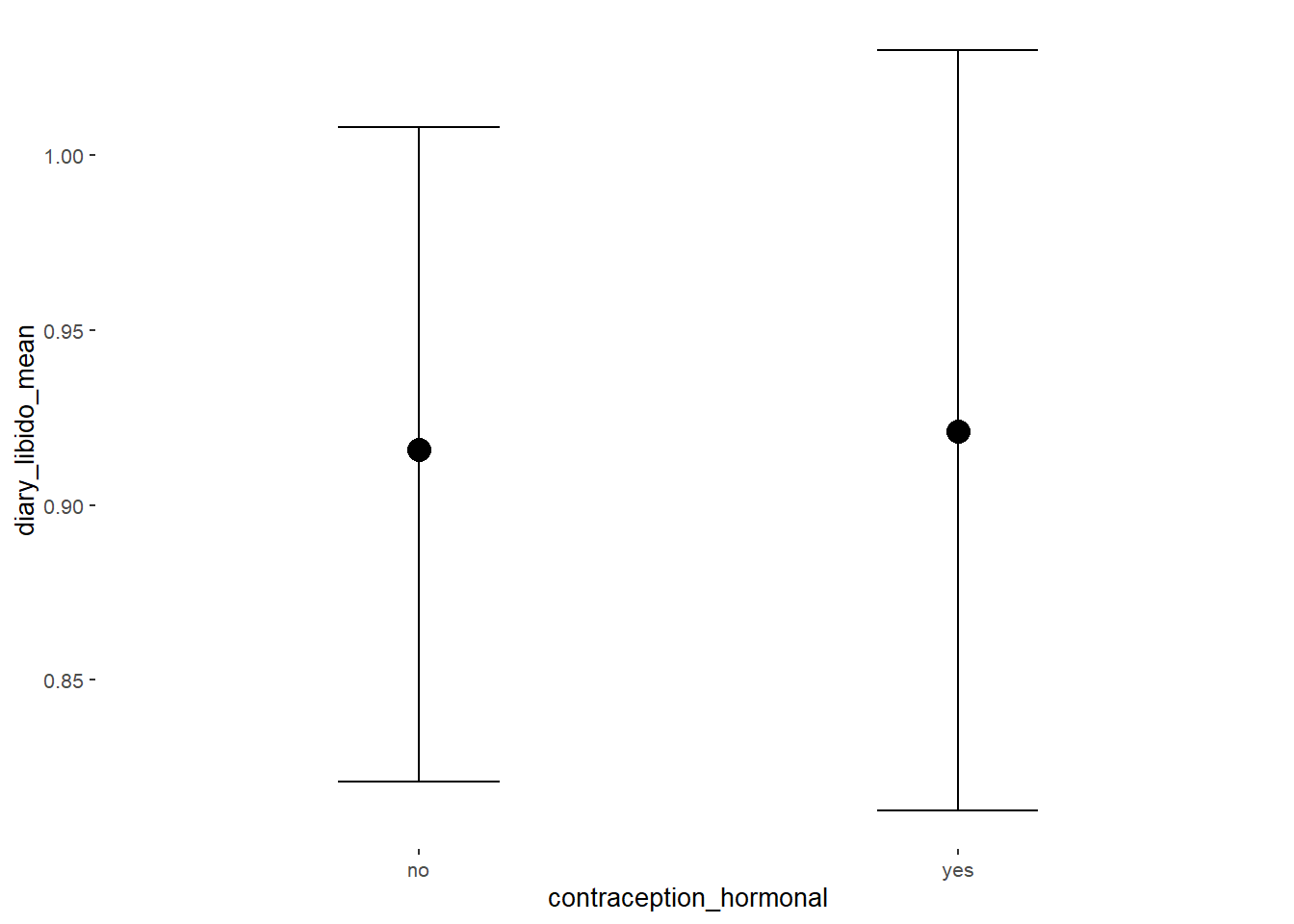
conditional_effects(m_hc_sexfreqpen,
effects = "contraception_hormonal",
conditions = data.frame(number_of_days = 1))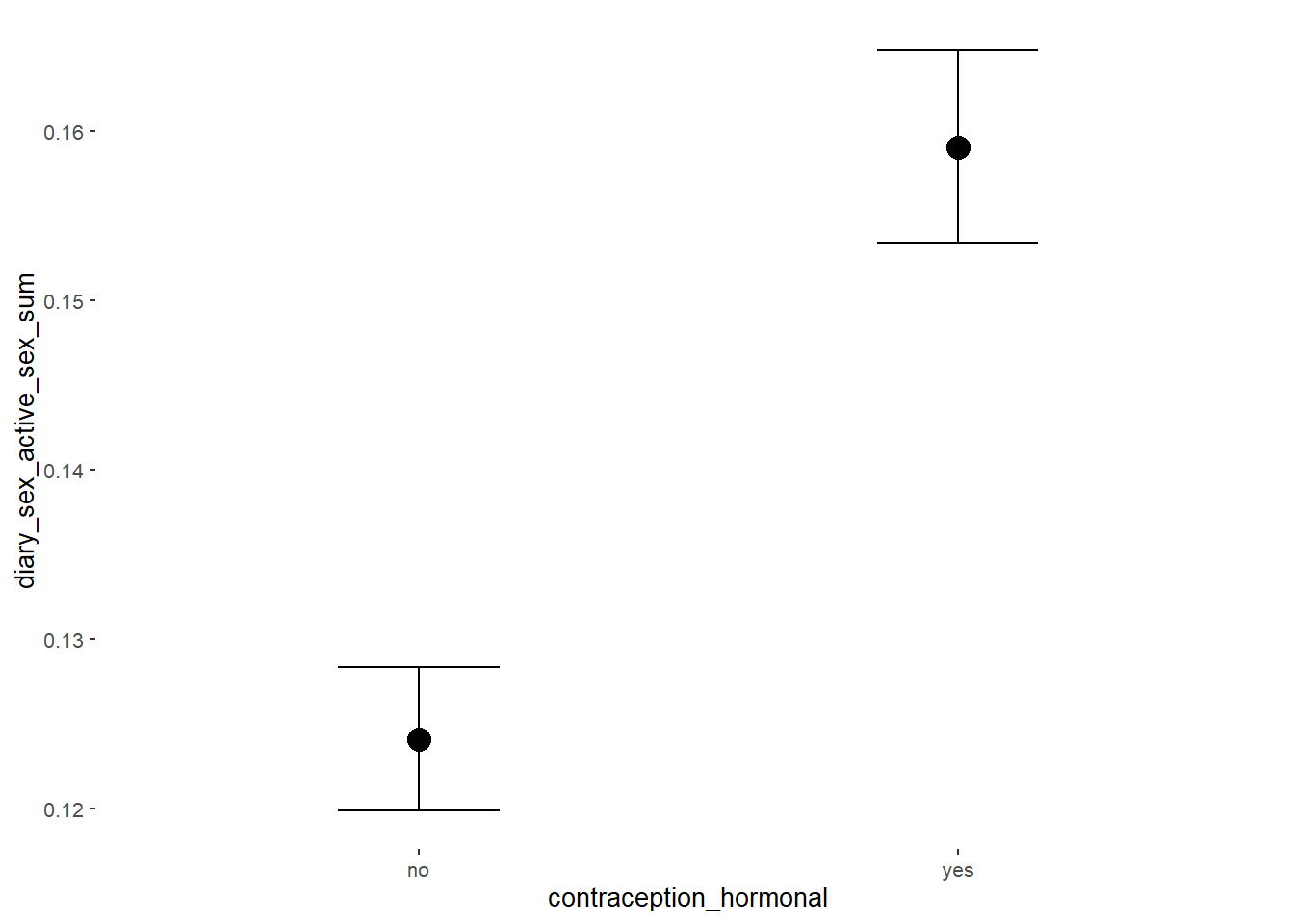
Forest Plot for Effect Sizes
m_hc_sexfreqpen %>%
spread_draws(b_contraception_hormonalyes) %>%
pivot_longer(cols = c(b_contraception_hormonalyes),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_contraception_hormonalyes",
"Contraception", NA),
condition = ifelse(condition %contains% "b_contraception_hormonalyes",
"Hormonal Contraception", NA)) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.05))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.05, 0.05), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")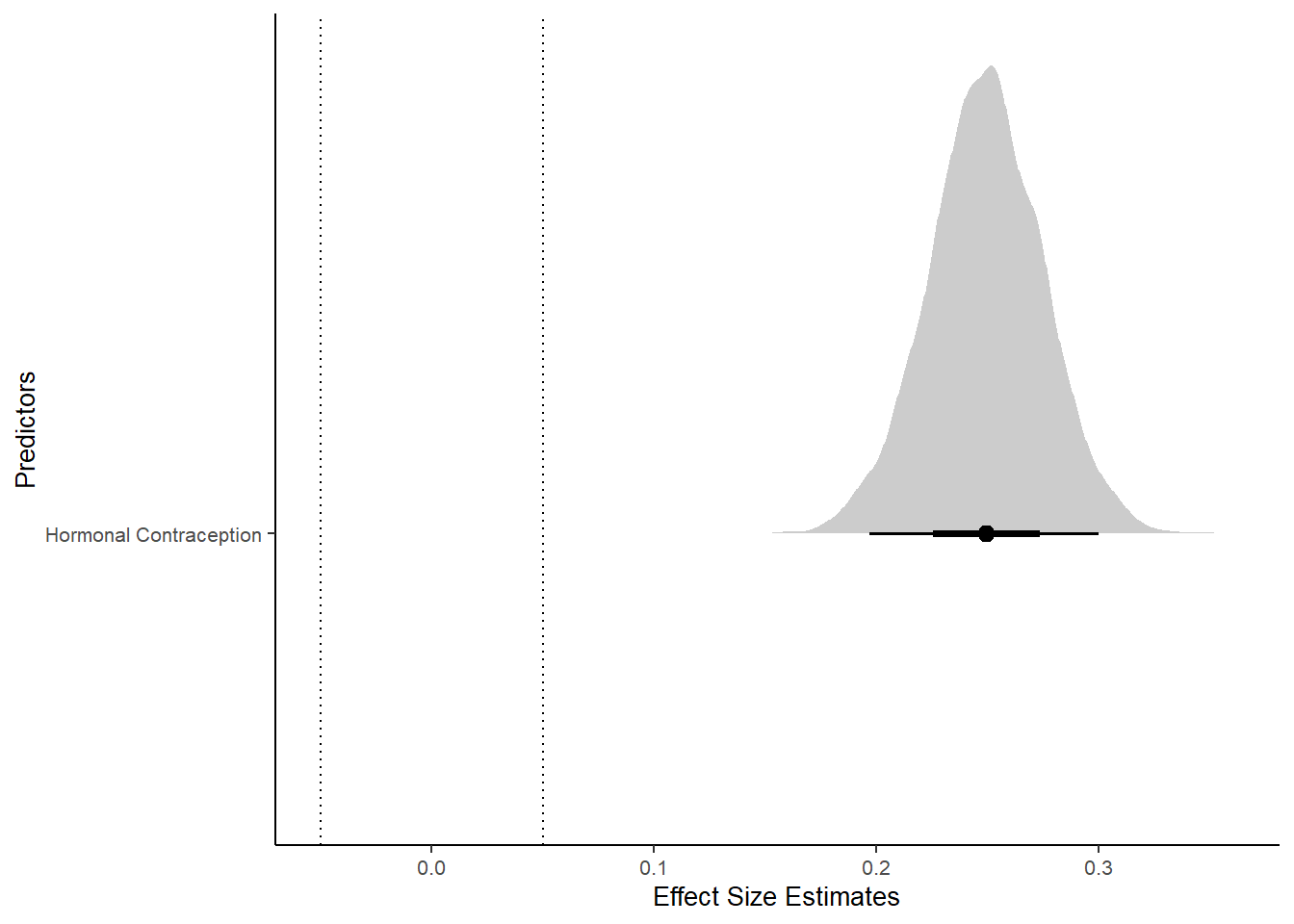
Masturbation Frequency
Model
Summary
## Family: poisson
## Links: mu = log
## Formula: diary_masturbation_sum ~ offset(log(number_of_days)) + contraception_hormonal
## Data: data (Number of observations: 897)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## Intercept -1.88 0.02 -1.90 -1.85 1.00 4566 3063
## contraception_hormonalyes -0.40 0.03 -0.44 -0.35 1.00 2847 2414
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_hc_masfreq, range = c(-0.05, 0.05), ci = 0.90,
parameters = "contraception"))## Picking joint bandwidth of 0.00367## Warning: Removed 399 rows containing non-finite values (stat_density_ridges).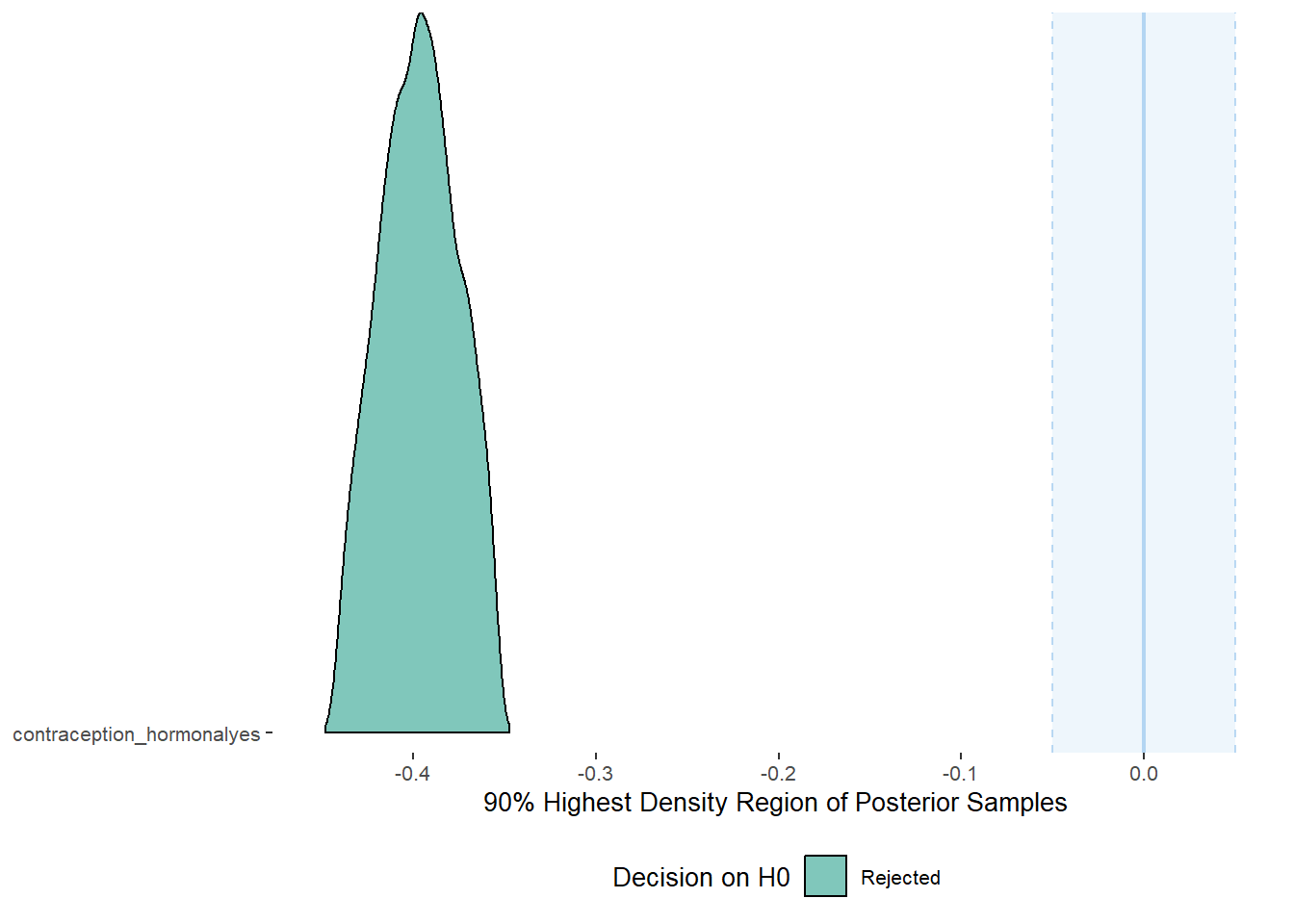
## # A tibble: 1 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.05 0.05 0 Rejected -0.441 -0.354 fixed conditio~Plots
conditional_effects(m_hc_masfreq,
effects = "contraception_hormonal",
conditions = data.frame(number_of_days = 1))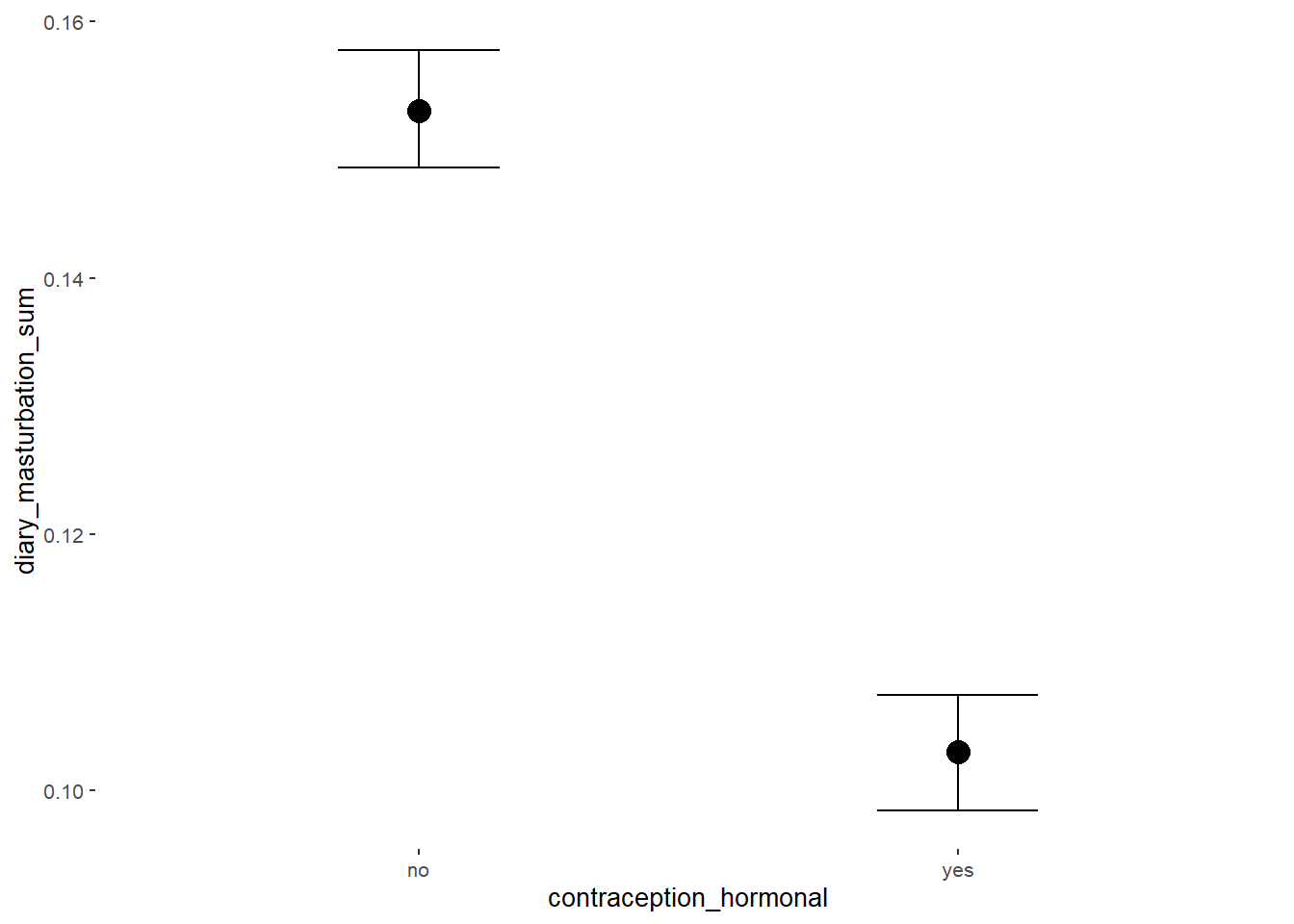
Forest Plot for Effect Sizes
m_hc_masfreq %>%
spread_draws(b_contraception_hormonalyes) %>%
pivot_longer(cols = c(b_contraception_hormonalyes),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_contraception_hormonalyes",
"Contraception", NA),
condition = ifelse(condition %contains% "b_contraception_hormonalyes",
"Hormonal Contraception", NA)) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.05))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.05, 0.05), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")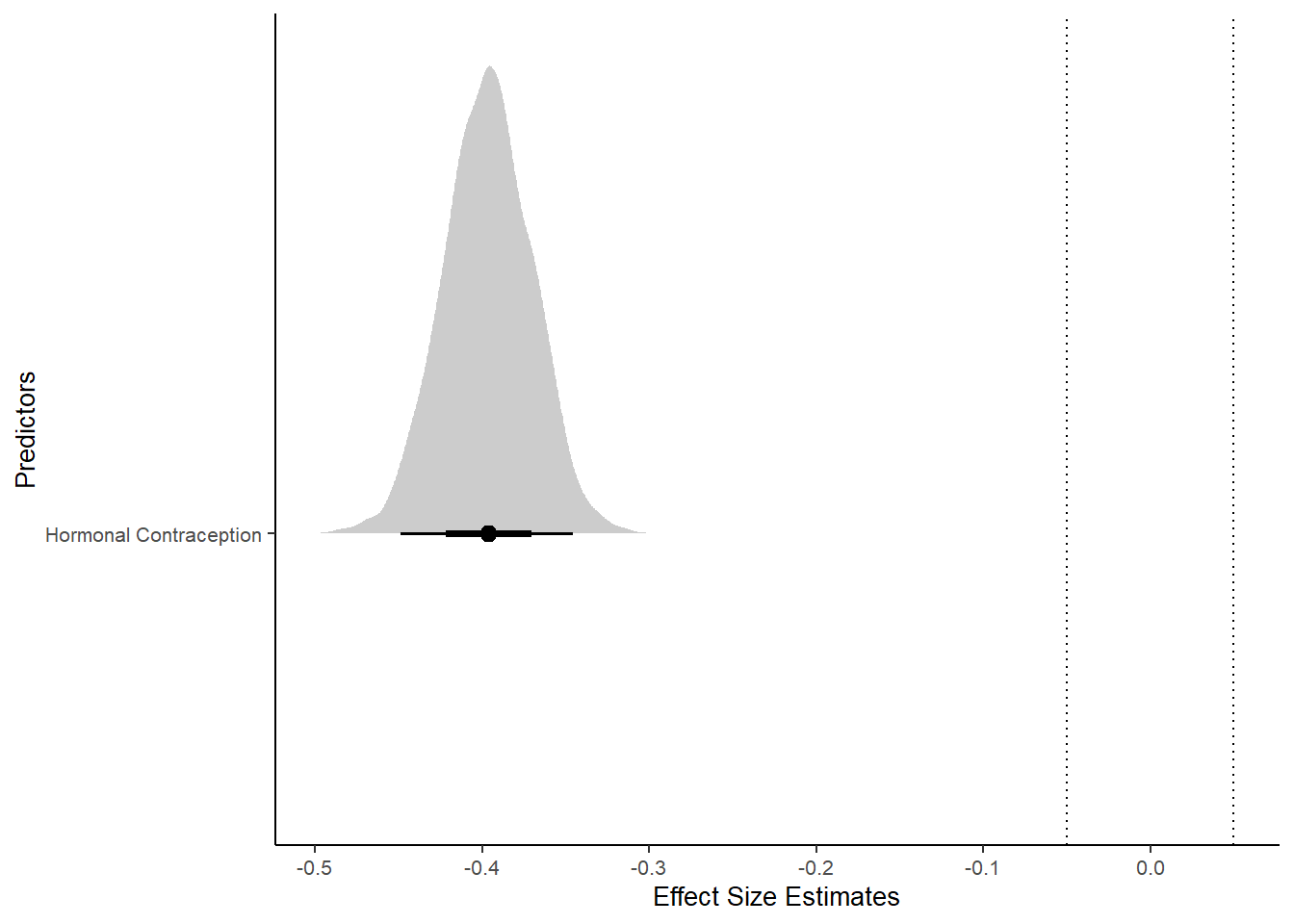
Controlled Models: Effects of Hormonal Contraceptives
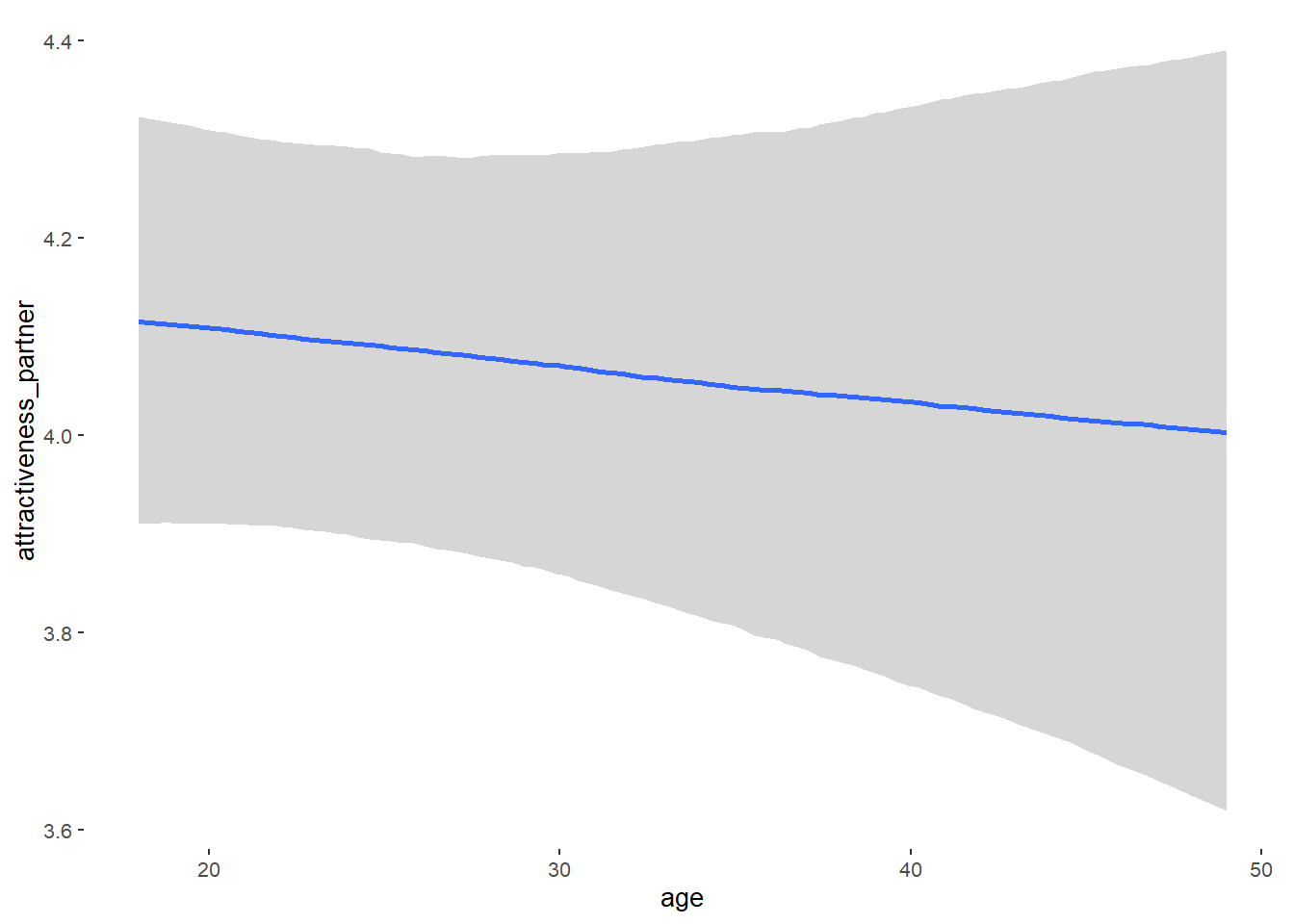
Attractiveness of Partner
Model
m_hc_atrr_controlled = brm(attractiveness_partner ~ contraception_hormonal +
age + net_income + relationship_duration_factor +
education_years +
bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open +
religiosity,
data = data, family = gaussian(),
file = "m_hc_atrr_controlled")## Warning: Rows containing NAs were excluded from the model.## Compiling Stan program...## Start sampling##
## SAMPLING FOR MODEL 'e2eceadeaf05fc7906f2a27d8d9a066f' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 0 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 10.787 seconds (Warm-up)
## Chain 1: 12.406 seconds (Sampling)
## Chain 1: 23.193 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL 'e2eceadeaf05fc7906f2a27d8d9a066f' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 0 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 10.786 seconds (Warm-up)
## Chain 2: 12.547 seconds (Sampling)
## Chain 2: 23.333 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL 'e2eceadeaf05fc7906f2a27d8d9a066f' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 0 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 10.674 seconds (Warm-up)
## Chain 3: 12.357 seconds (Sampling)
## Chain 3: 23.031 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL 'e2eceadeaf05fc7906f2a27d8d9a066f' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 0 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 10.736 seconds (Warm-up)
## Chain 4: 12.141 seconds (Sampling)
## Chain 4: 22.877 seconds (Total)
## Chain 4:## Warning: There were 4000 transitions after warmup that exceeded the maximum treedepth. Increase max_treedepth above 10. See
## http://mc-stan.org/misc/warnings.html#maximum-treedepth-exceeded## Warning: Examine the pairs() plot to diagnose sampling problems## Warning: The largest R-hat is 3.51, indicating chains have not mixed.
## Running the chains for more iterations may help. See
## http://mc-stan.org/misc/warnings.html#r-hat## Warning: Bulk Effective Samples Size (ESS) is too low, indicating posterior means and medians may be unreliable.
## Running the chains for more iterations may help. See
## http://mc-stan.org/misc/warnings.html#bulk-ess## Warning: Tail Effective Samples Size (ESS) is too low, indicating posterior variances and tail quantiles may be unreliable.
## Running the chains for more iterations may help. See
## http://mc-stan.org/misc/warnings.html#tail-essSummary
## Warning: Parts of the model have not converged (some Rhats are > 1.05). Be careful when analysing the results!
## We recommend running more iterations and/or setting stronger priors.## Family: gaussian
## Links: mu = identity; sigma = identity
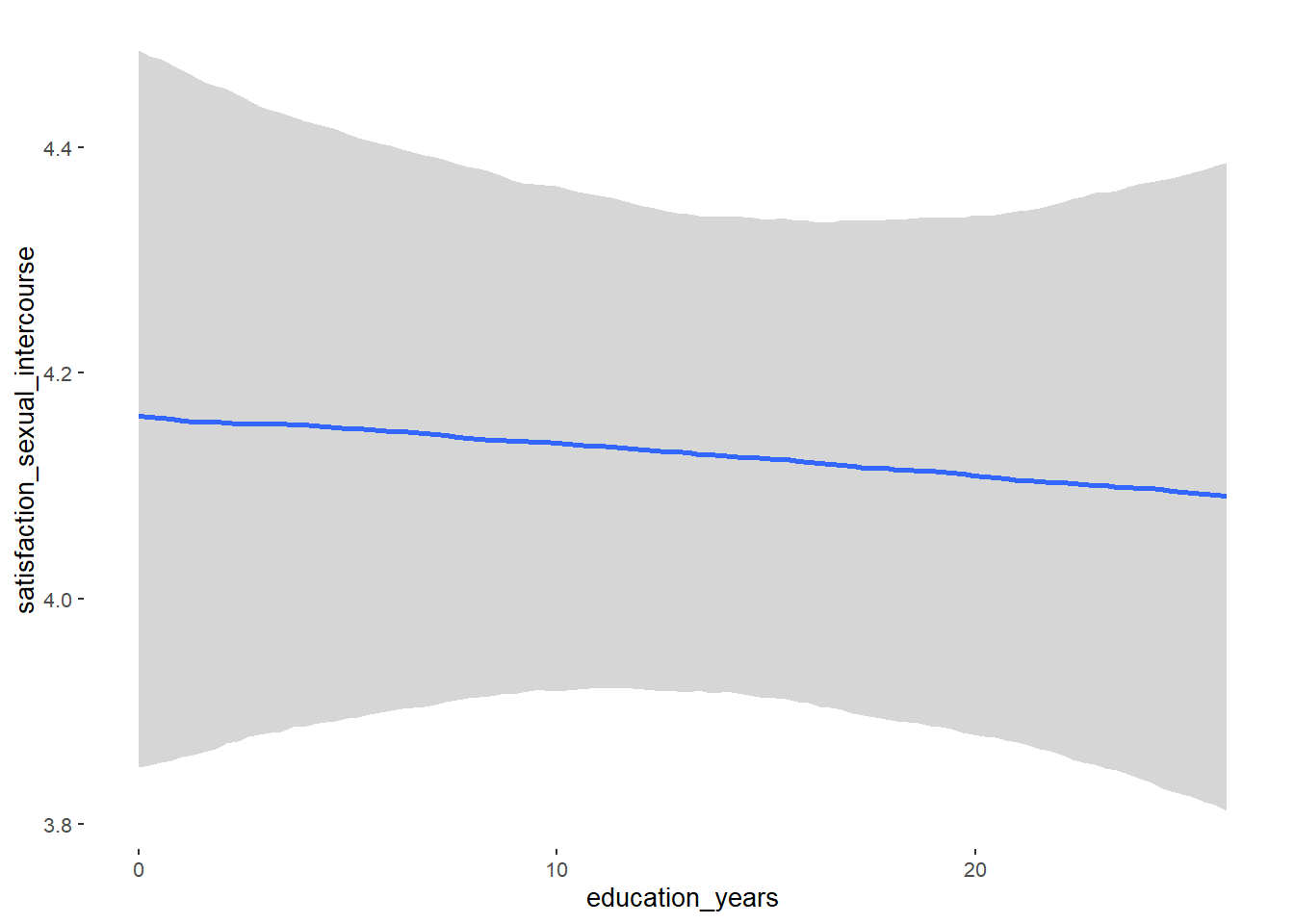
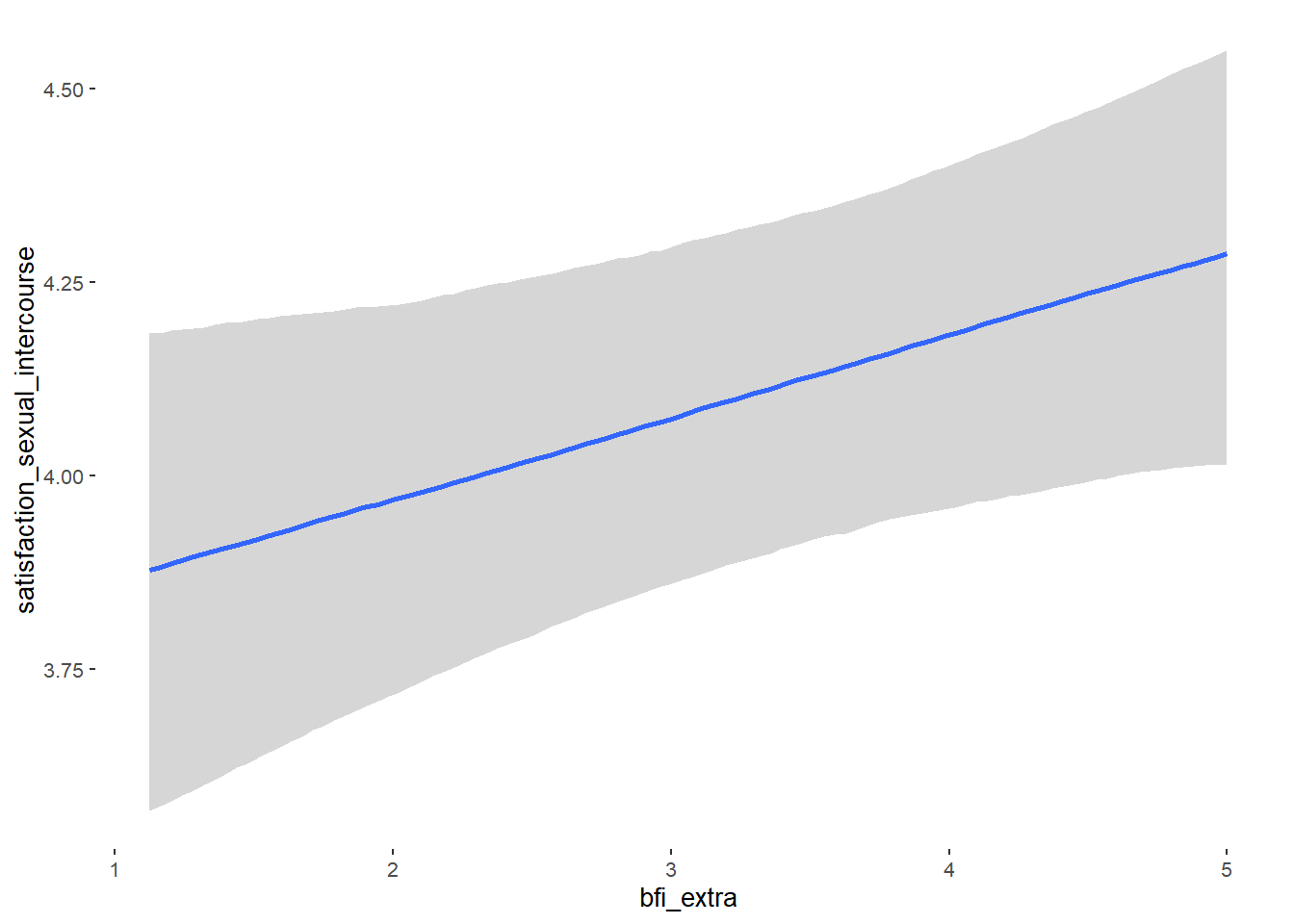
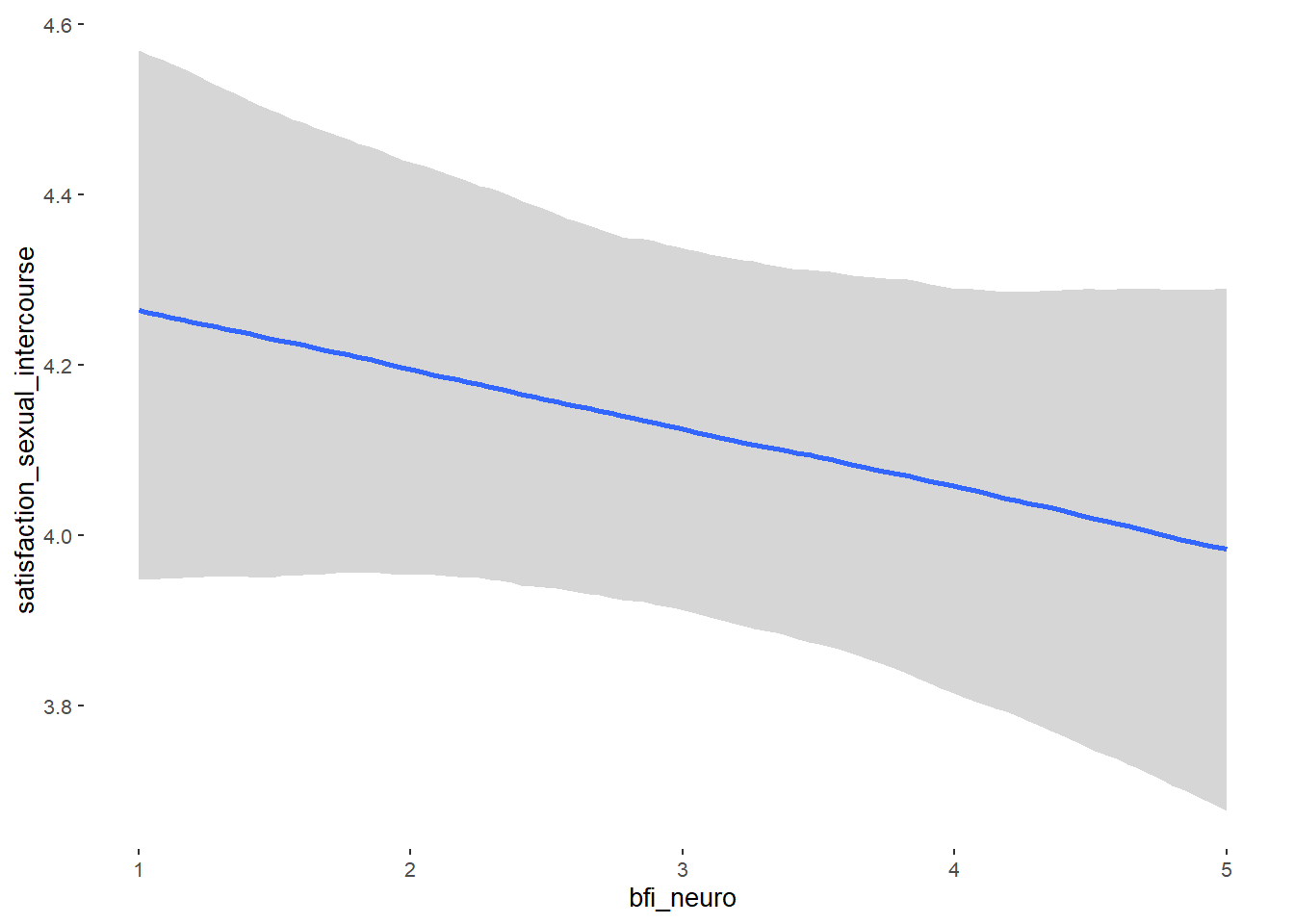
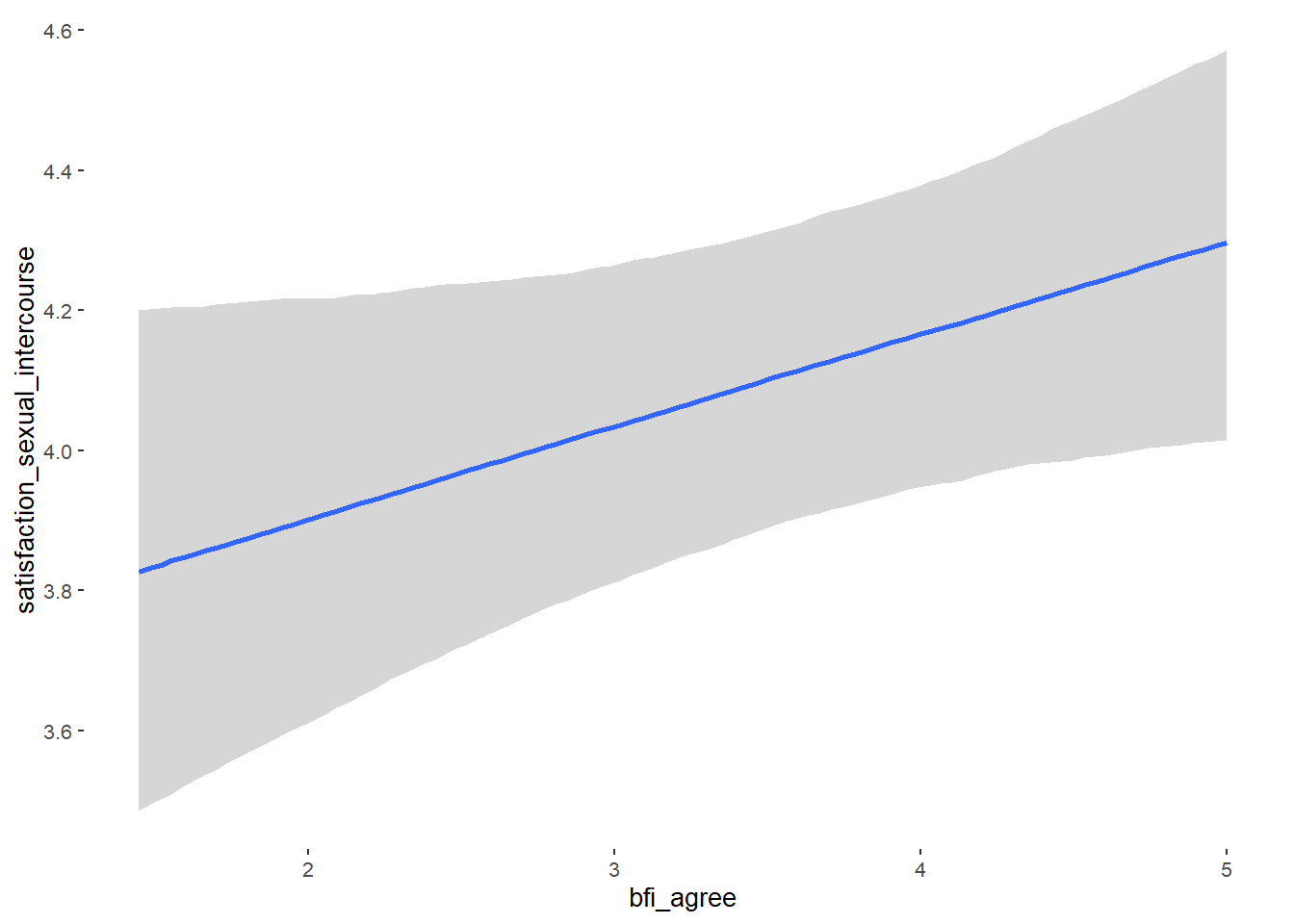
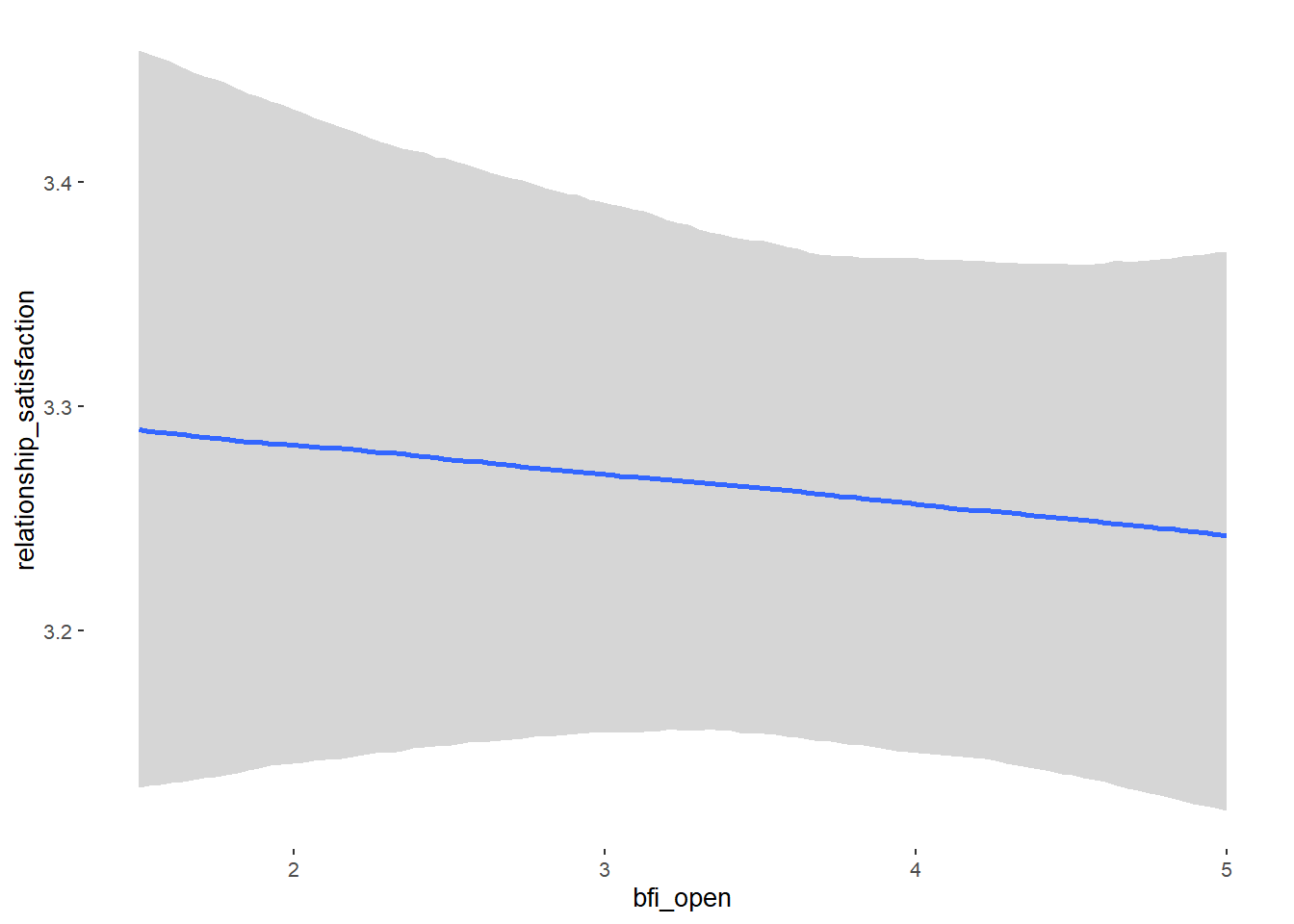
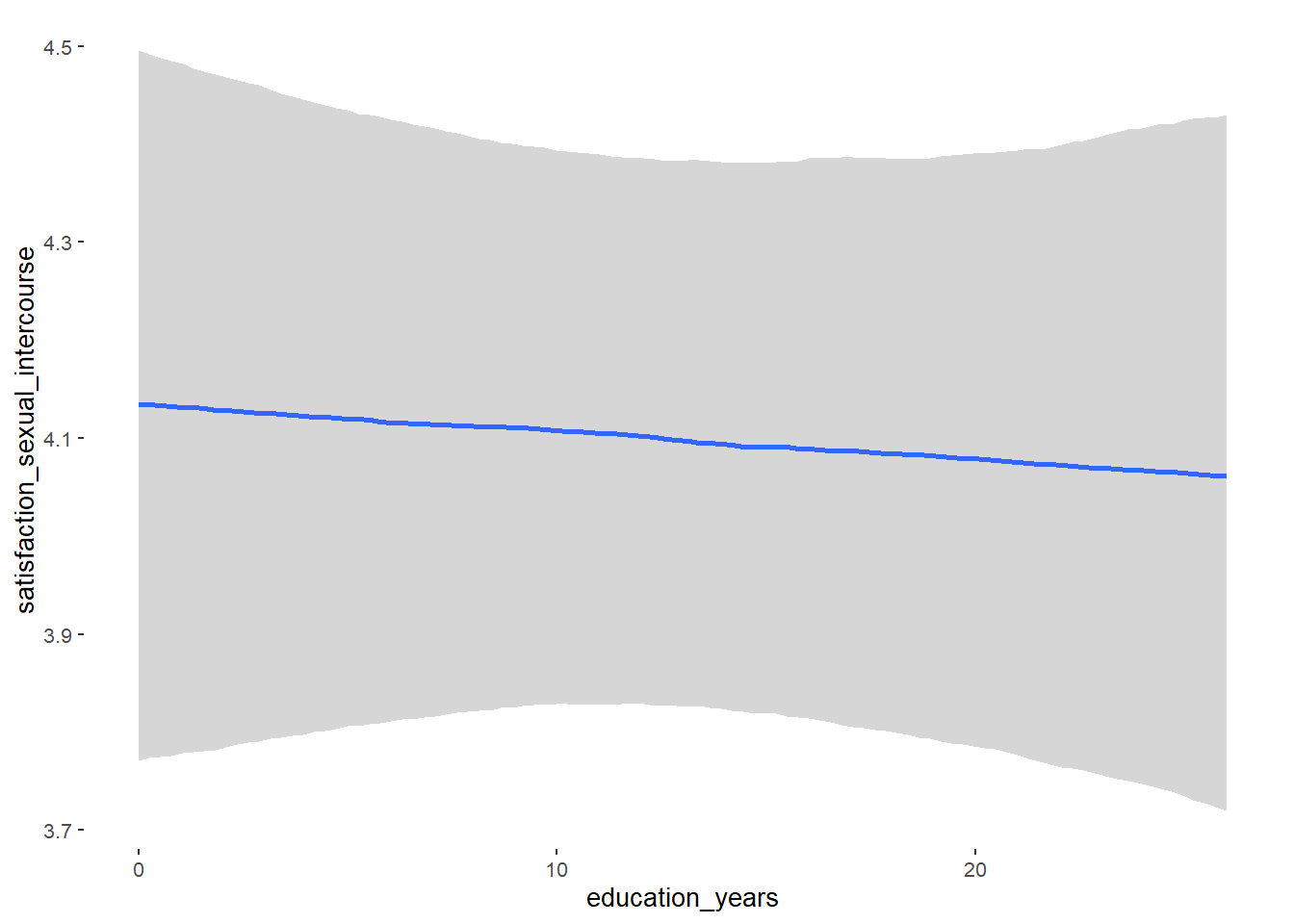
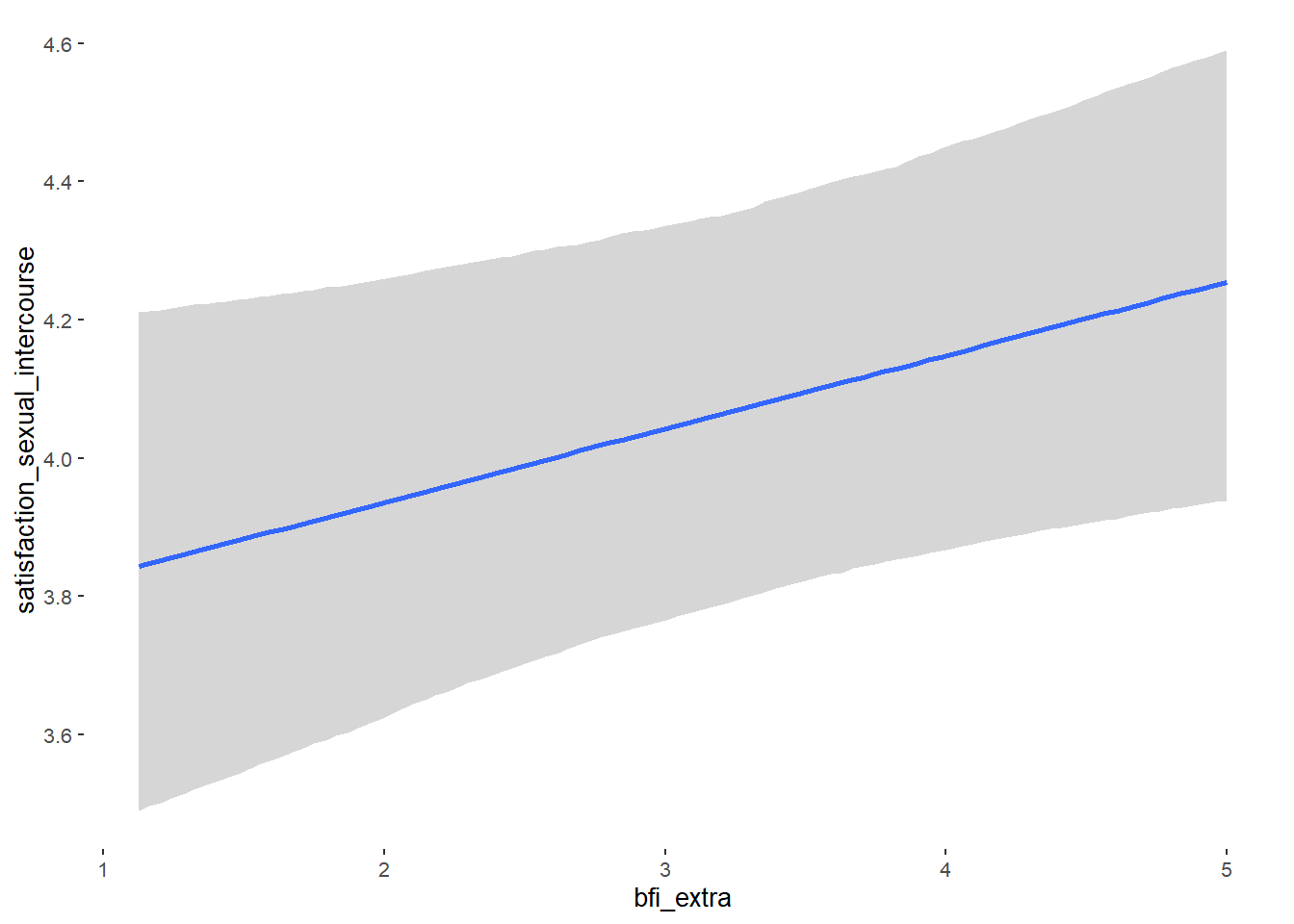
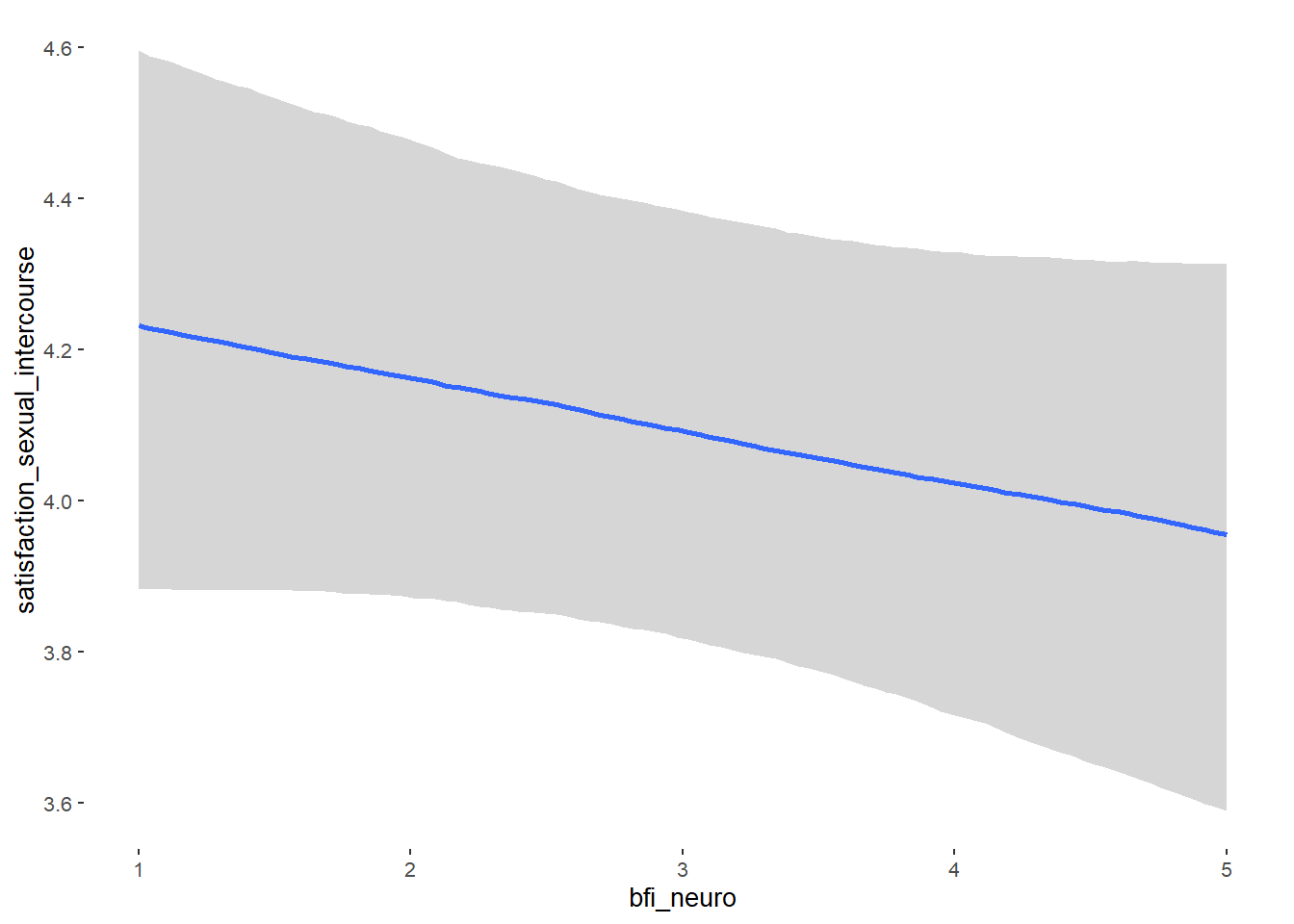
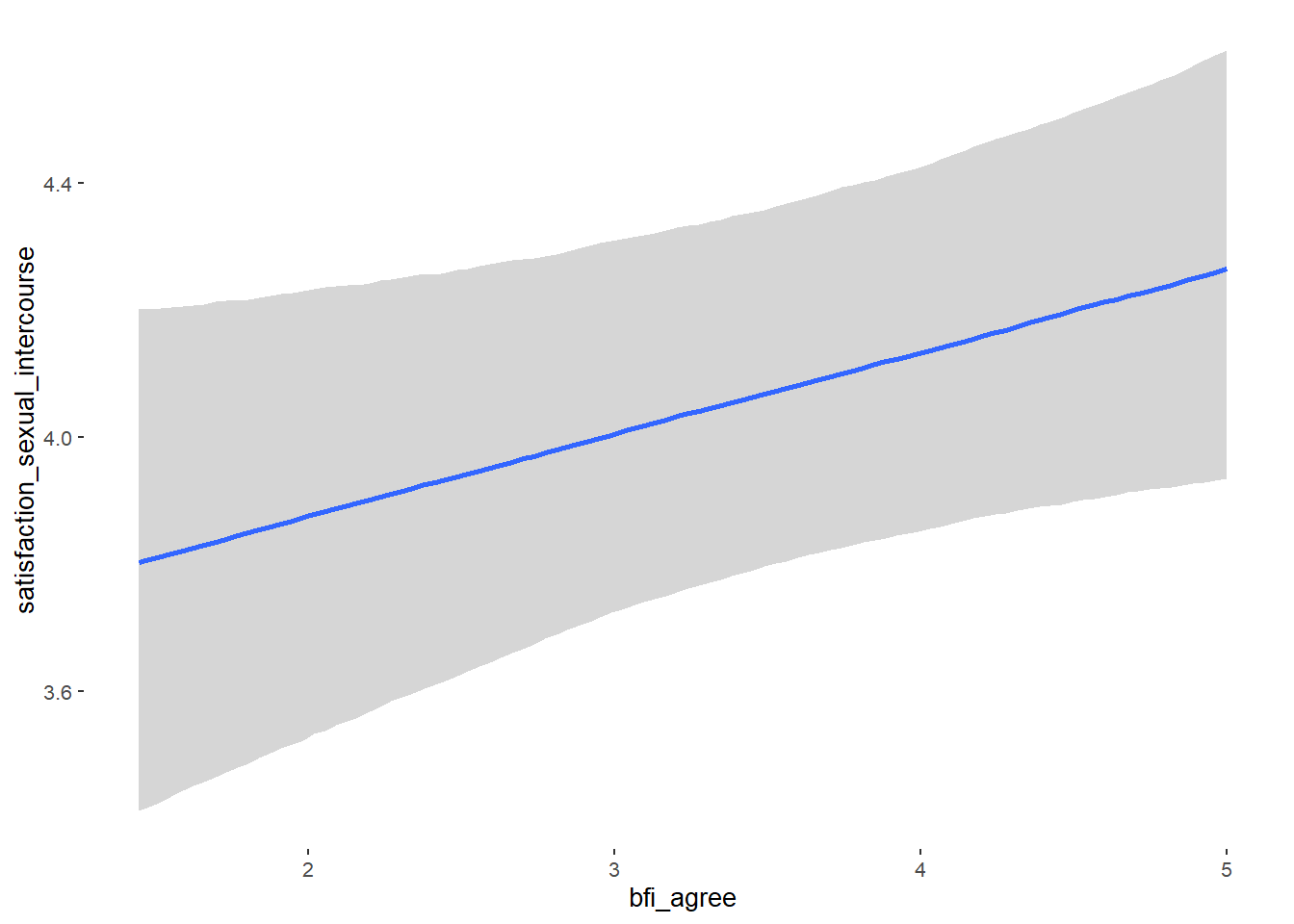
## Formula: attractiveness_partner ~ contraception_hormonal + age + net_income + relationship_duration_factor + education_years + bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open + religiosity
## Data: data (Number of observations: 774)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept 302.50 600.94 -565.96 1209.77 3.51 4
## contraception_hormonalyes 0.07 0.05 -0.02 0.14 1.09 36
## age -0.00 0.01 -0.01 0.01 1.07 62
## net_incomeeuro_500_1000 0.04 0.05 -0.04 0.13 1.06 62
## net_incomeeuro_1000_2000 0.13 0.07 0.03 0.25 1.06 58
## net_incomeeuro_2000_3000 0.18 0.11 -0.01 0.34 1.05 51
## net_incomeeuro_gt_3000 0.21 0.20 -0.13 0.56 1.04 59
## net_incomedont_tell -0.01 0.16 -0.25 0.27 1.04 78
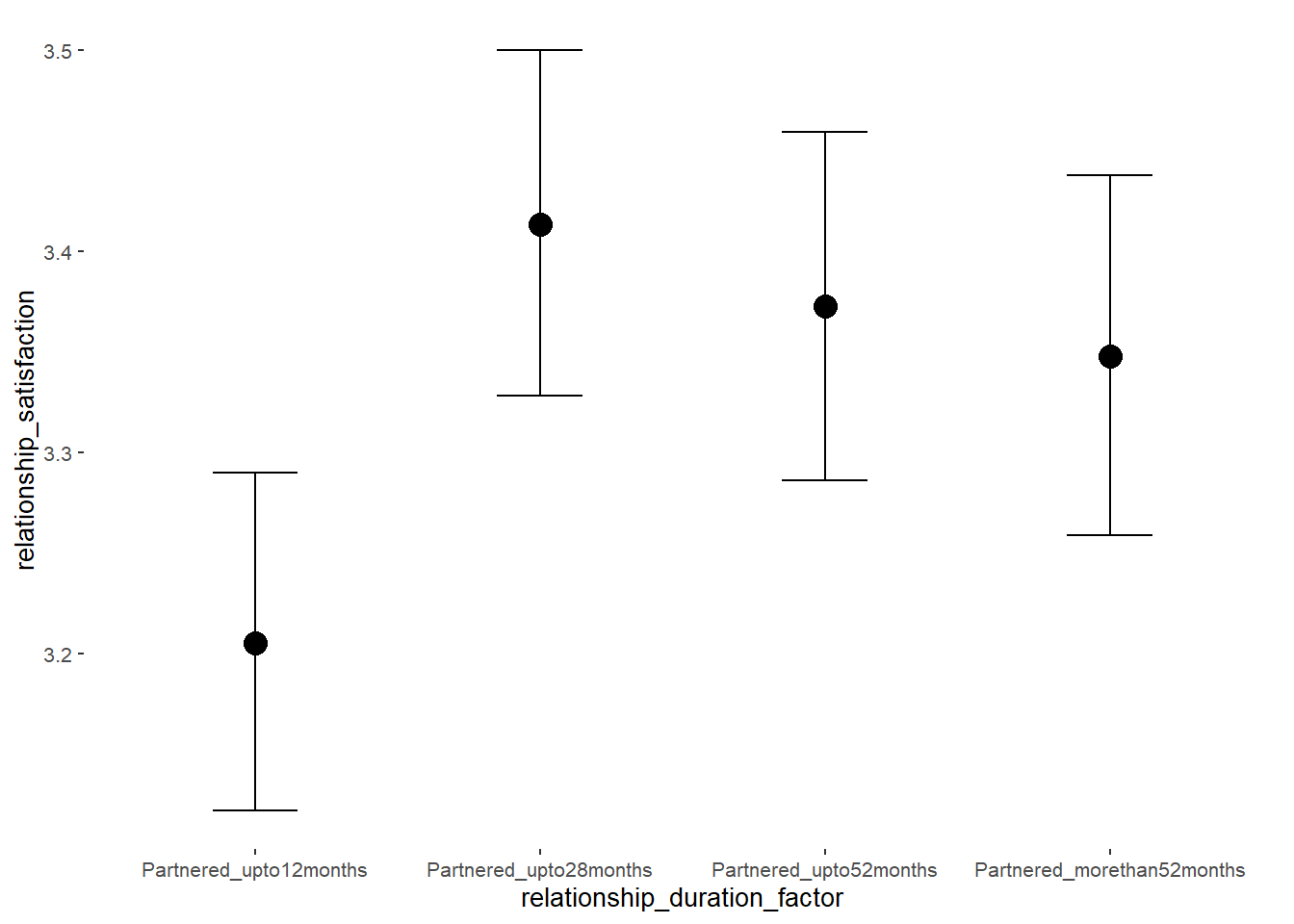
## relationship_duration_factorPartnered_upto12months -299.19 600.90 -1206.68 569.31 3.51 4
## relationship_duration_factorPartnered_upto28months -299.08 600.90 -1206.47 569.33 3.51 4
## relationship_duration_factorPartnered_upto52months -299.24 600.90 -1206.62 569.31 3.51 4
## relationship_duration_factorPartnered_morethan52months -299.34 600.89 -1206.73 569.15 3.51 4
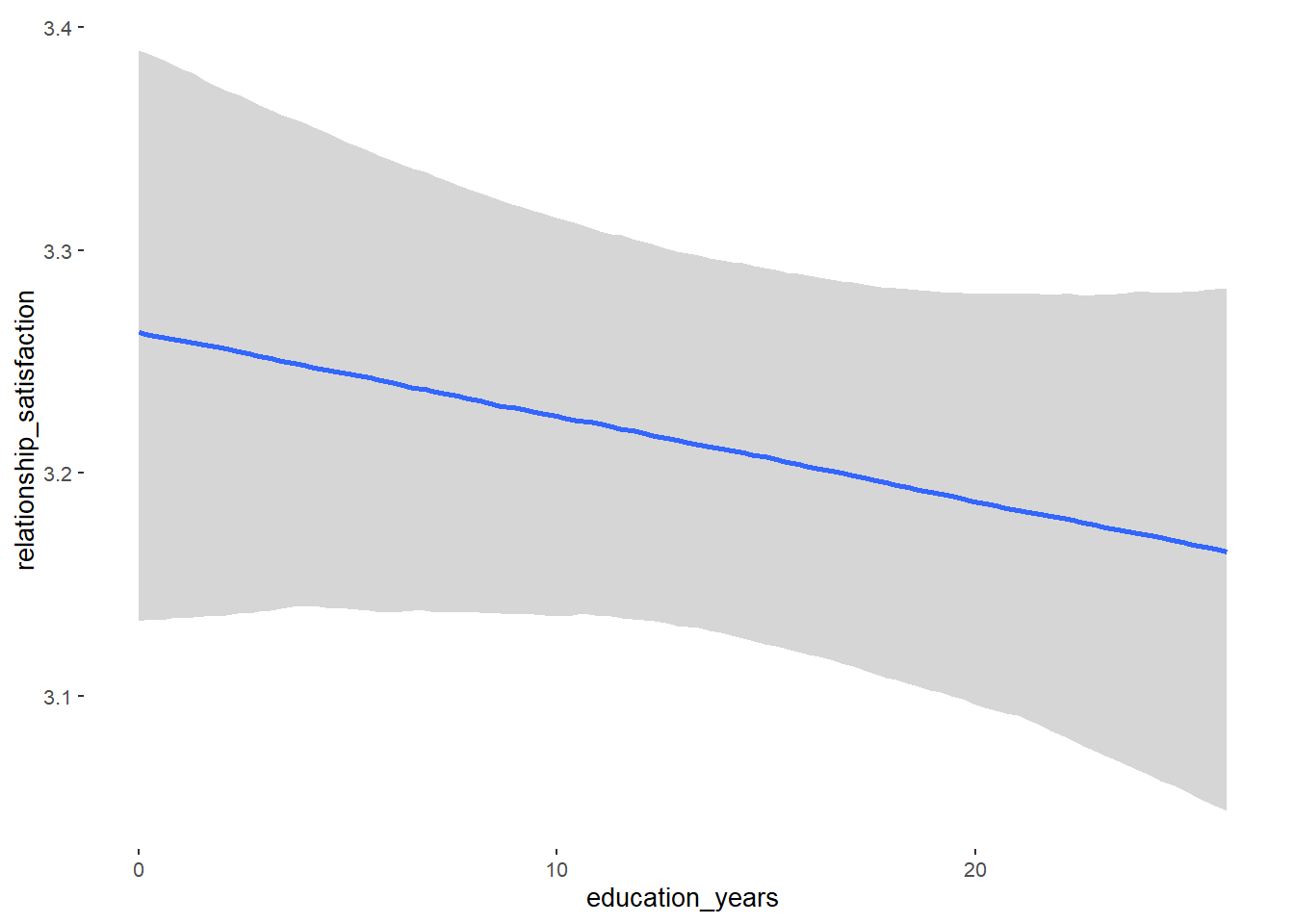
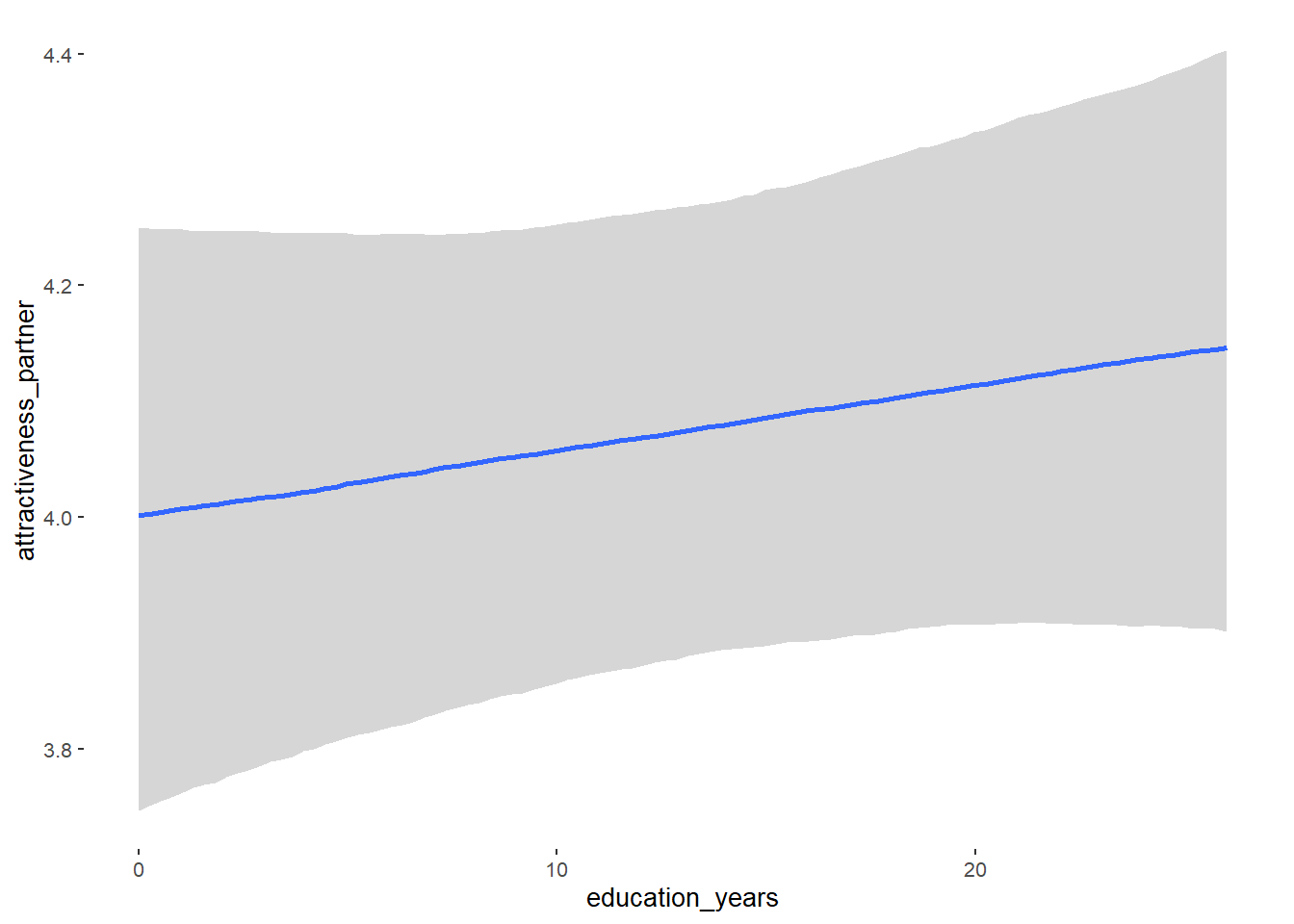
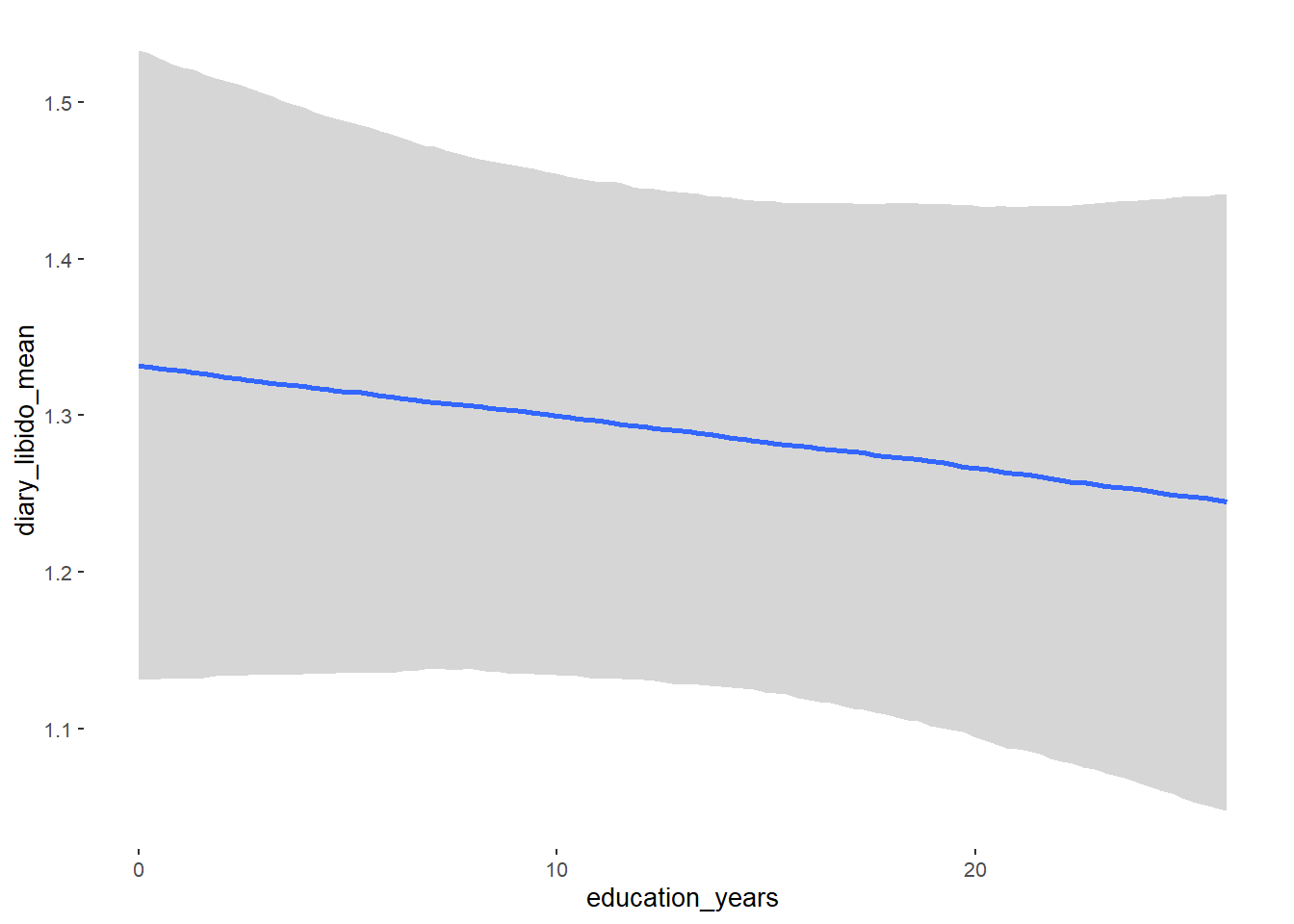
## education_years 0.01 0.01 -0.00 0.02 1.05 59
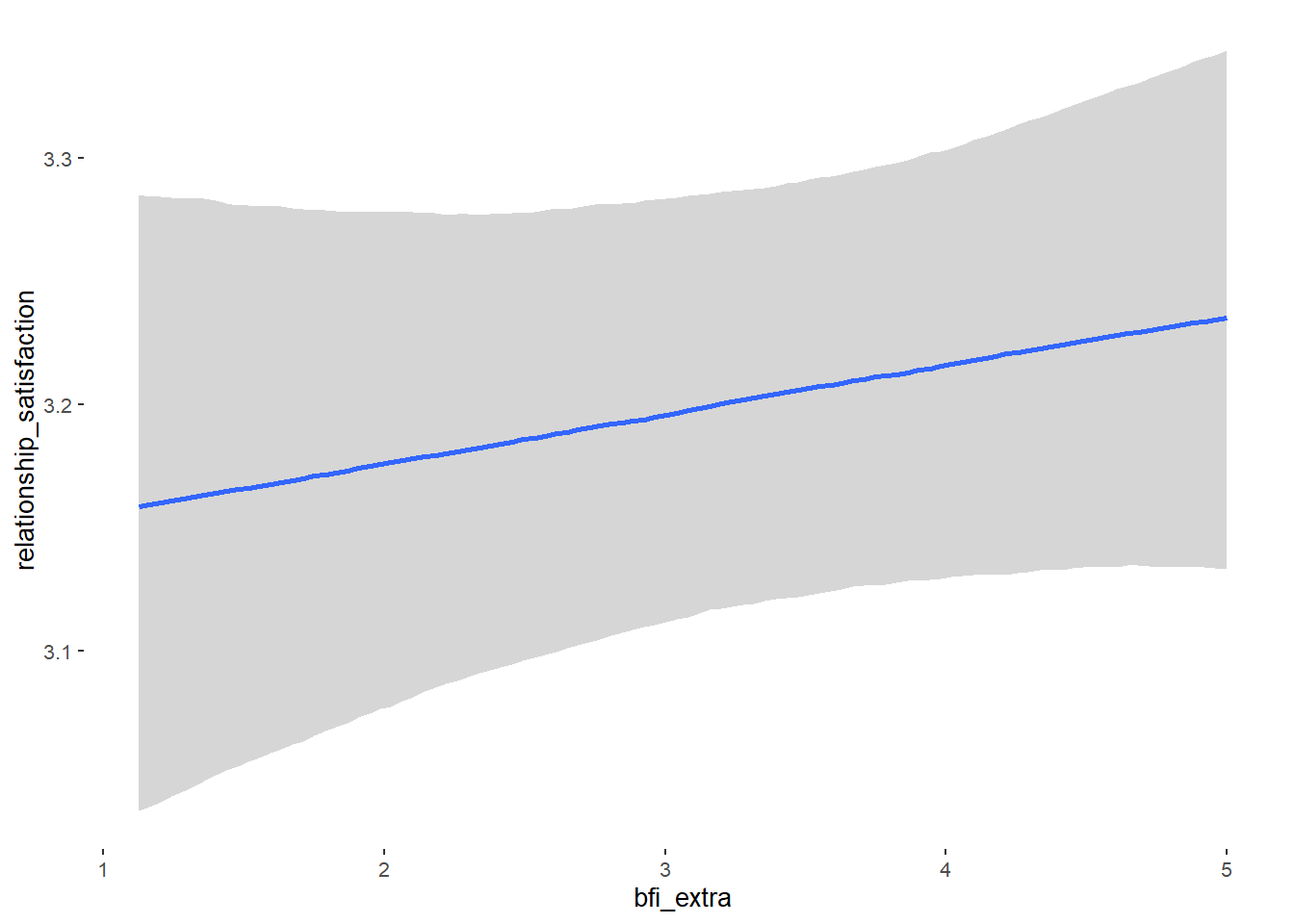
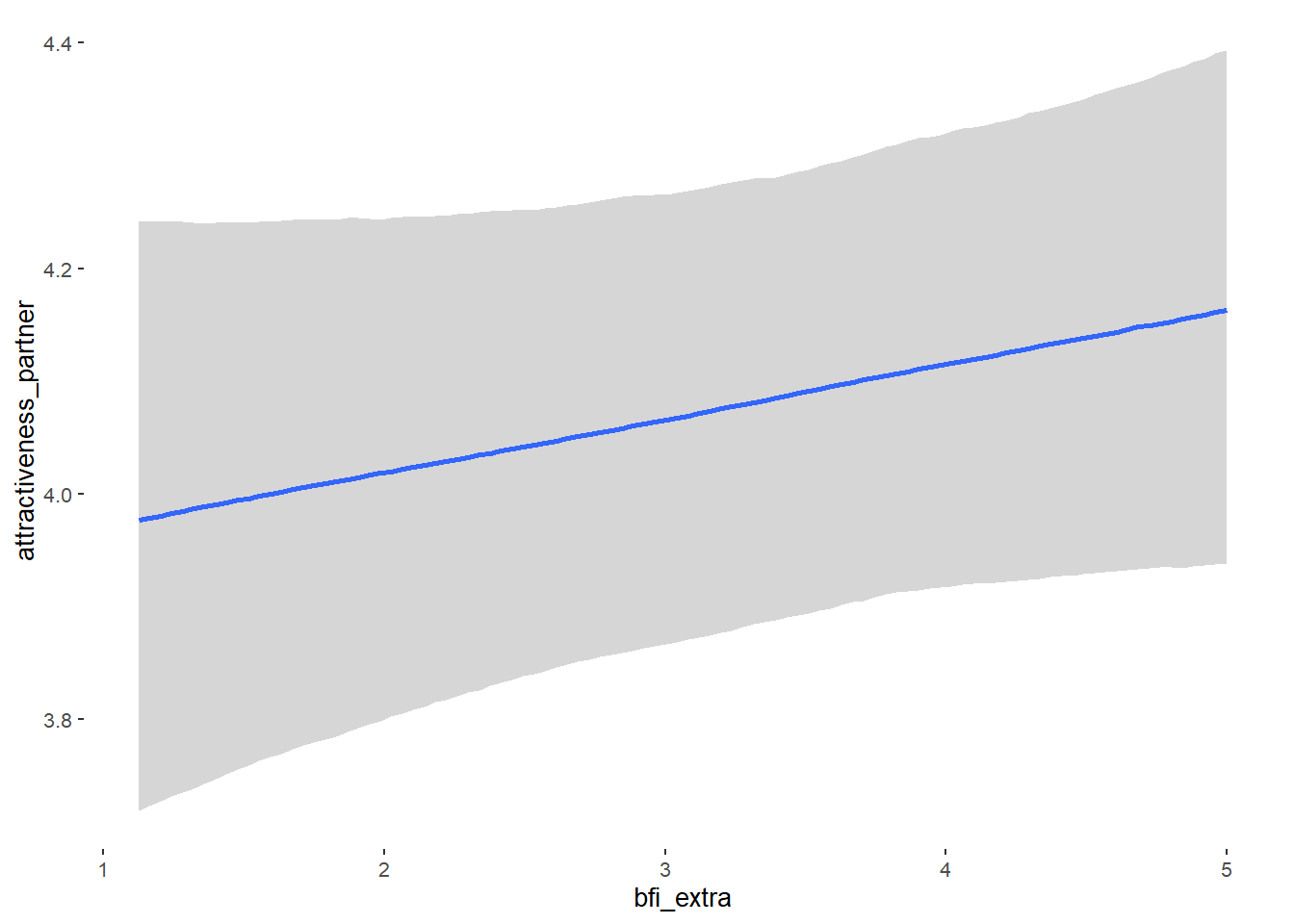
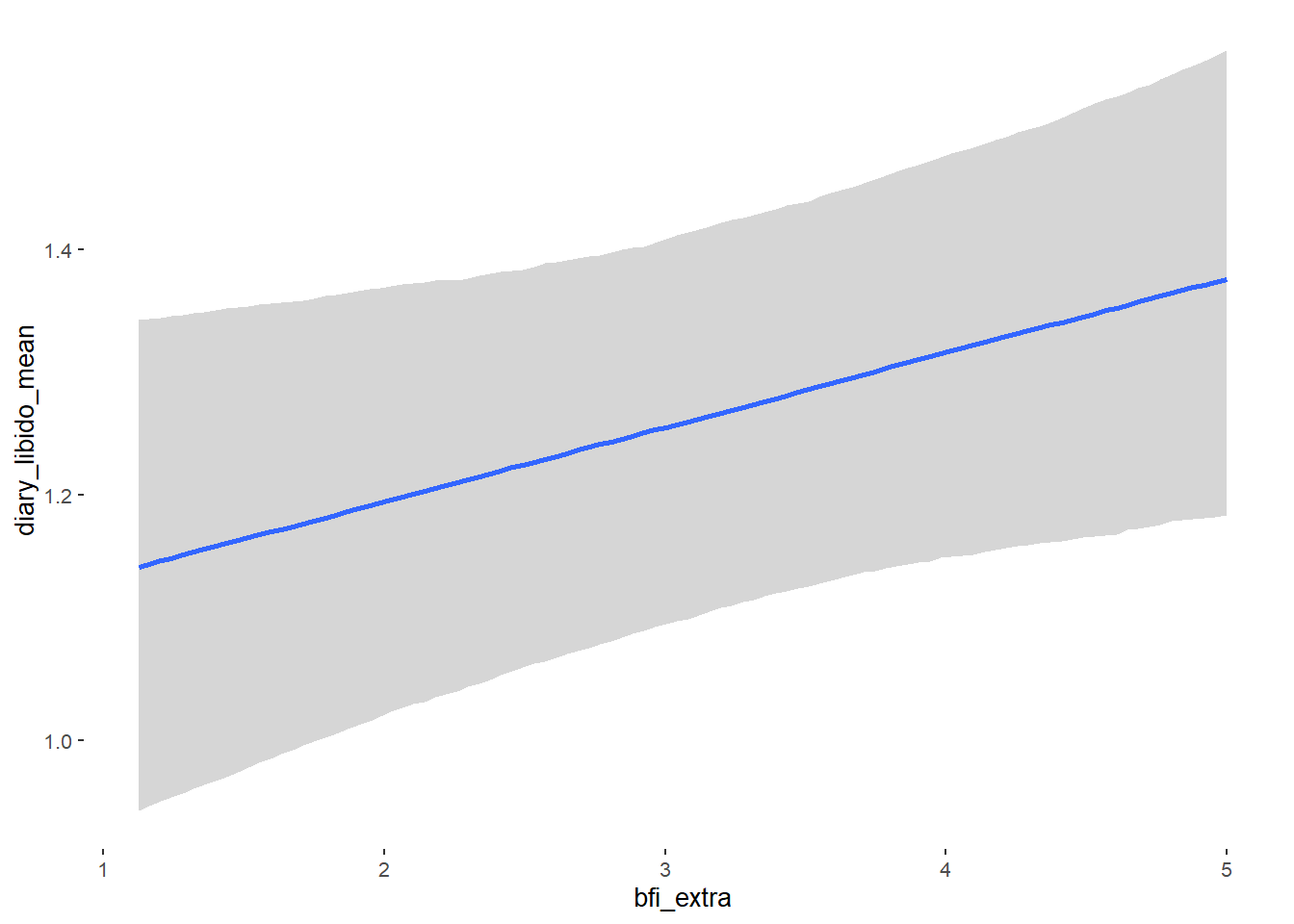
## bfi_extra 0.05 0.03 -0.01 0.10 1.10 38
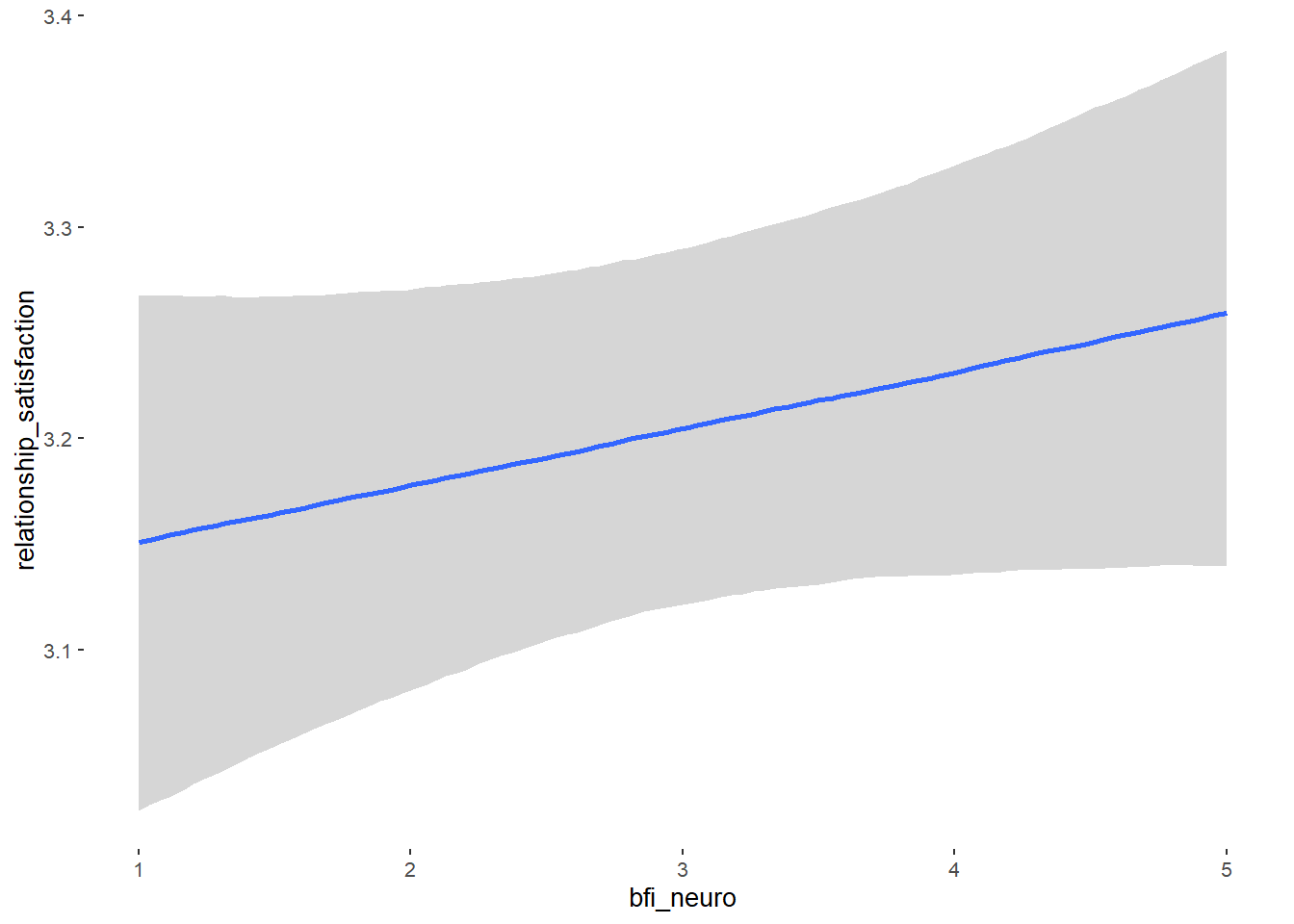
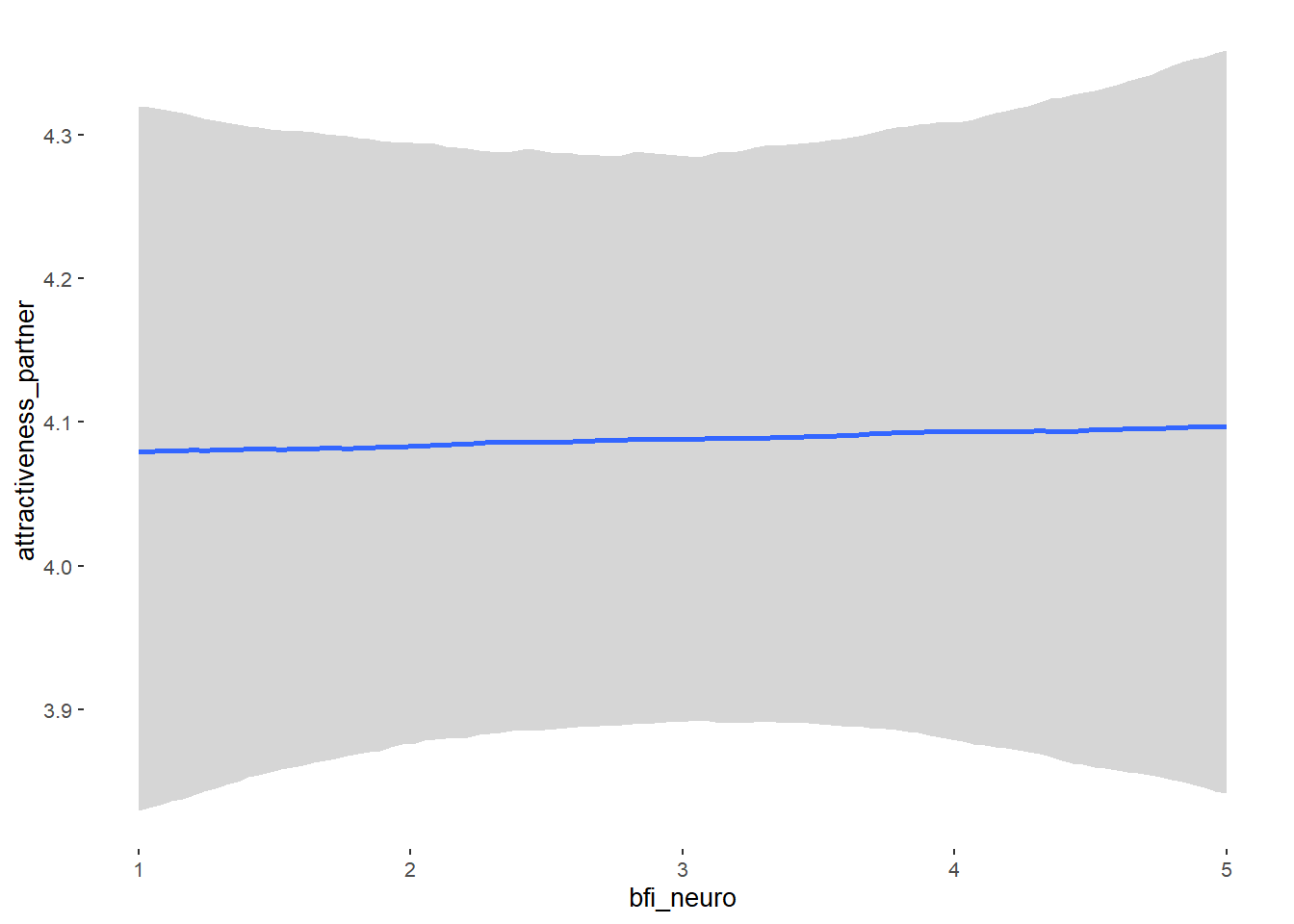
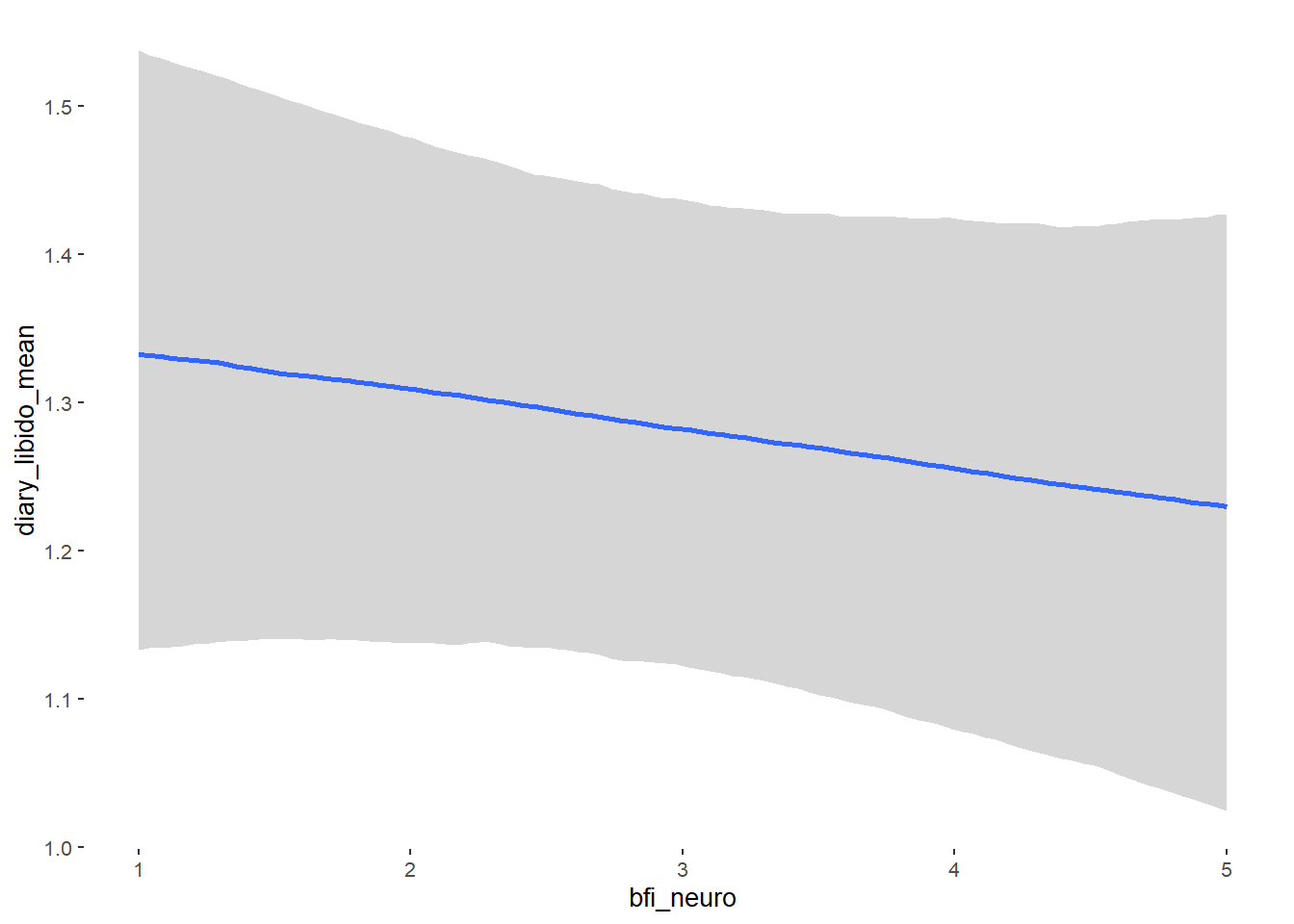
## bfi_neuro -0.00 0.04 -0.06 0.06 1.04 61
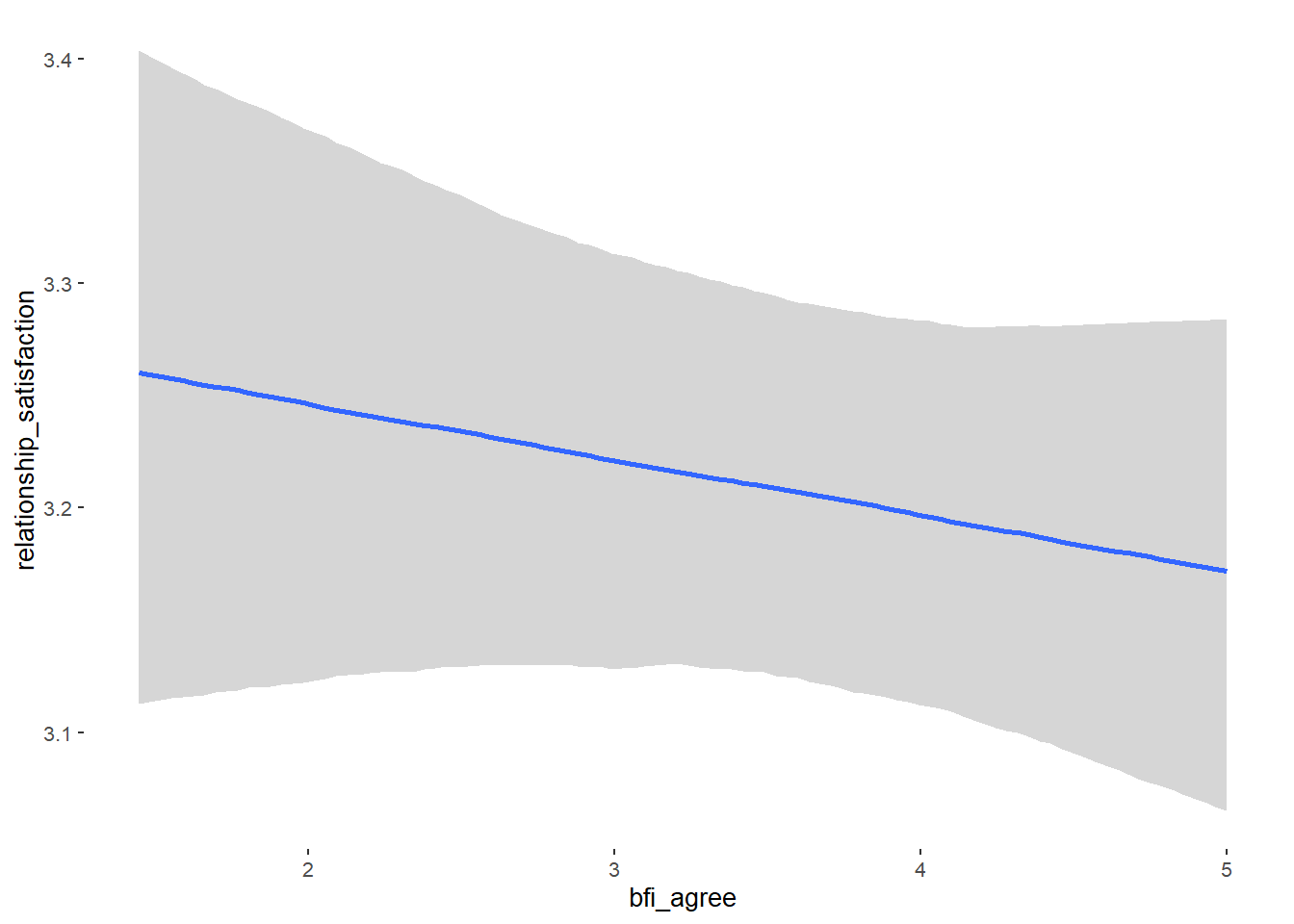
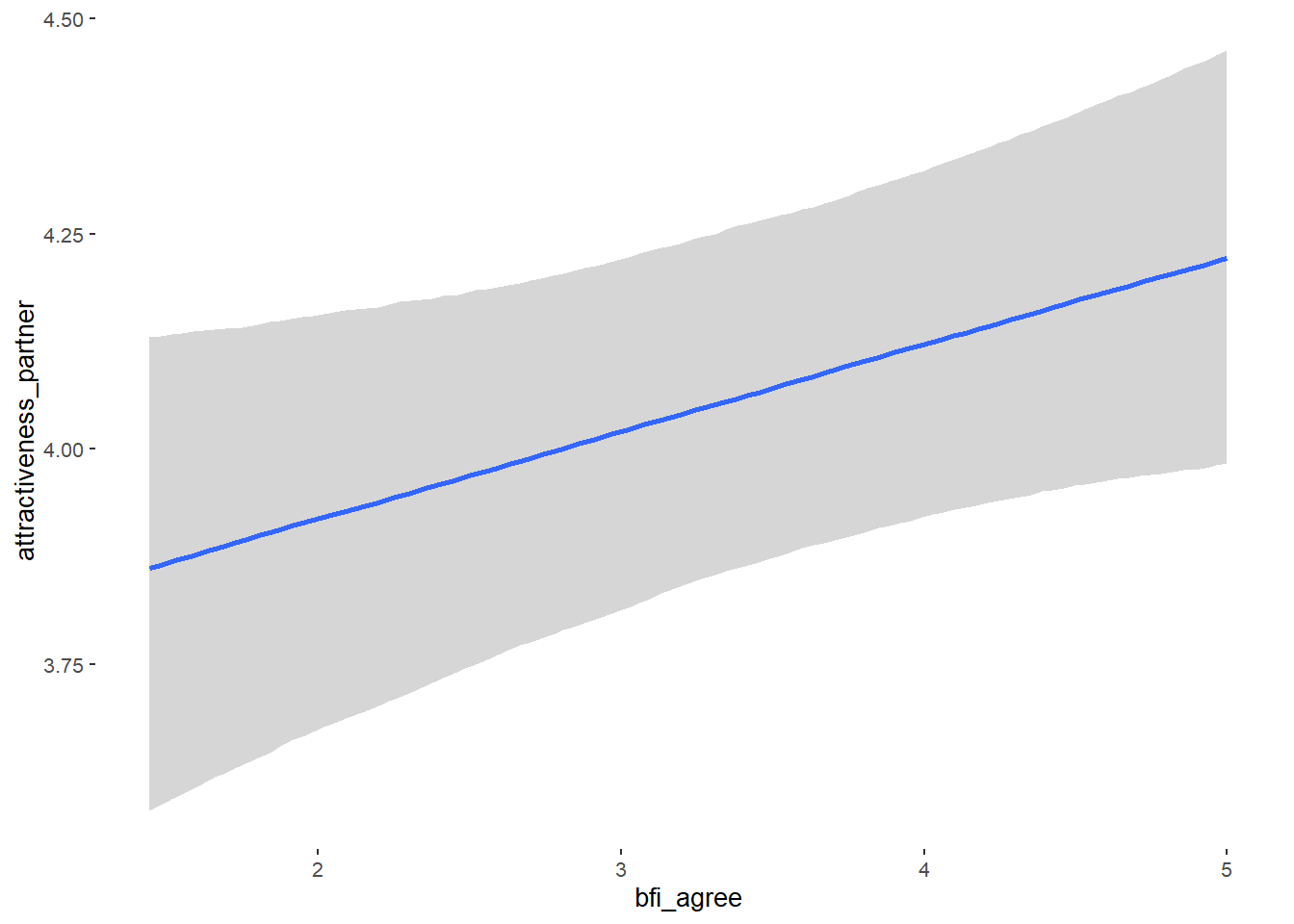
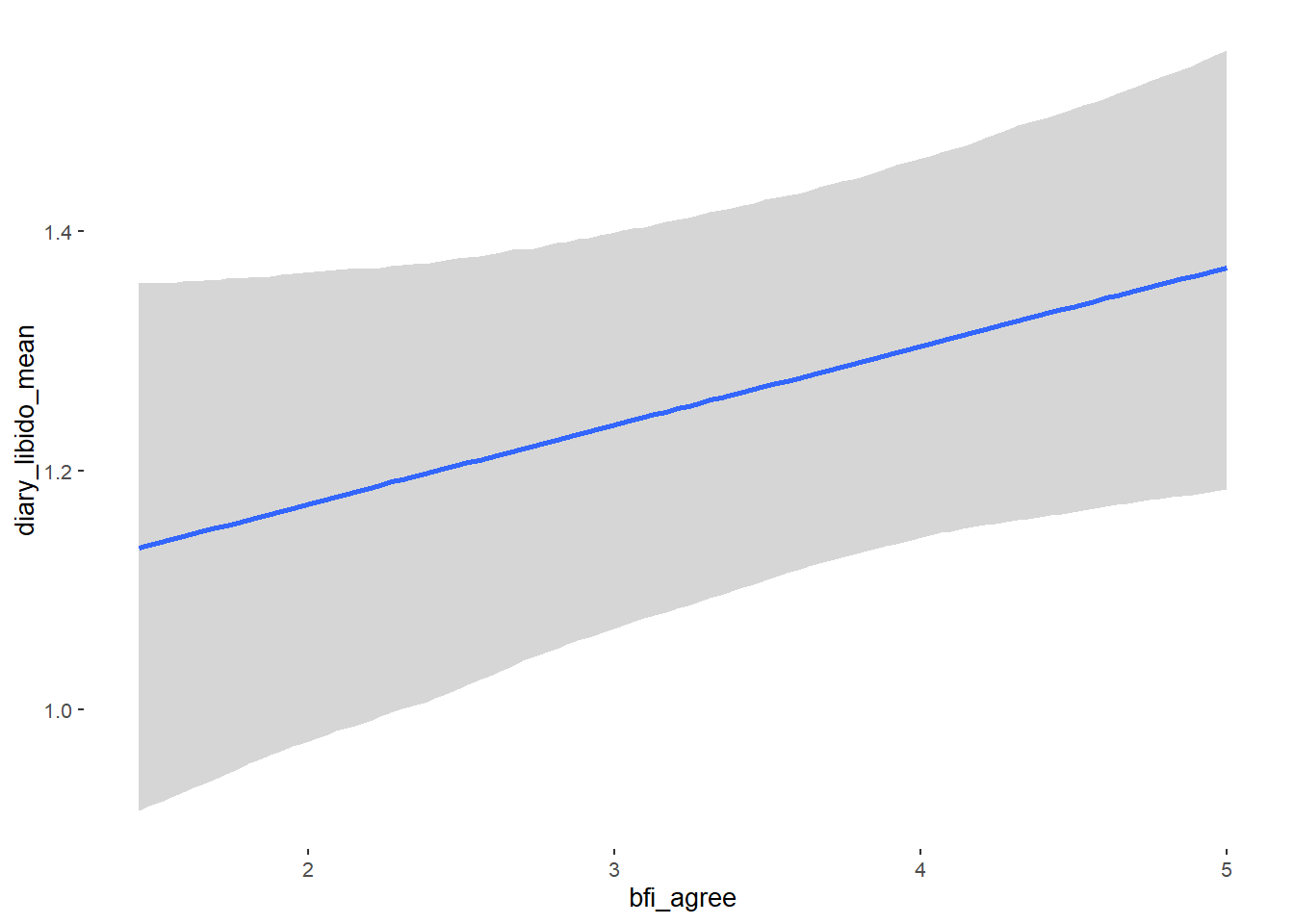
## bfi_agree 0.09 0.05 0.02 0.17 1.15 21
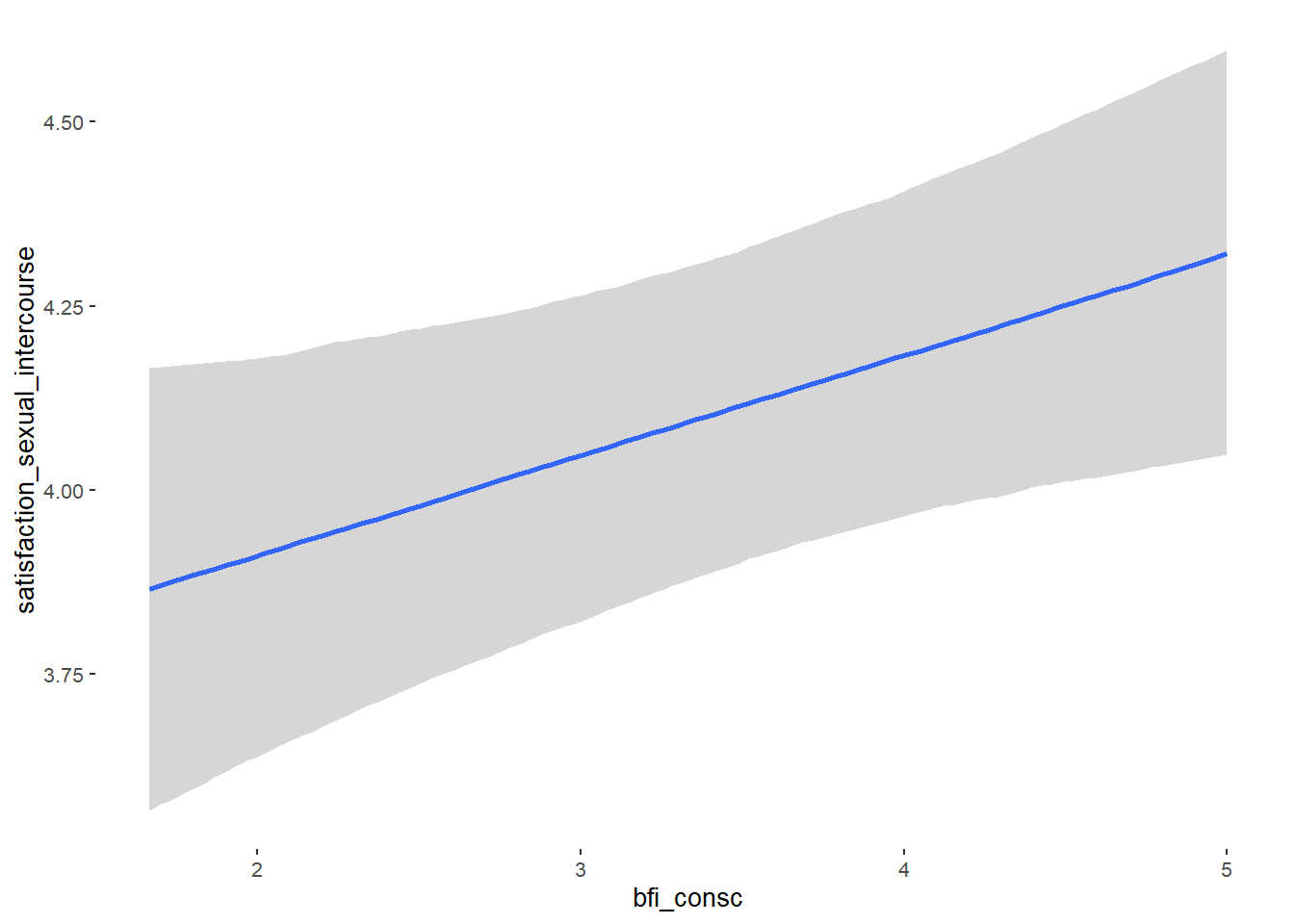
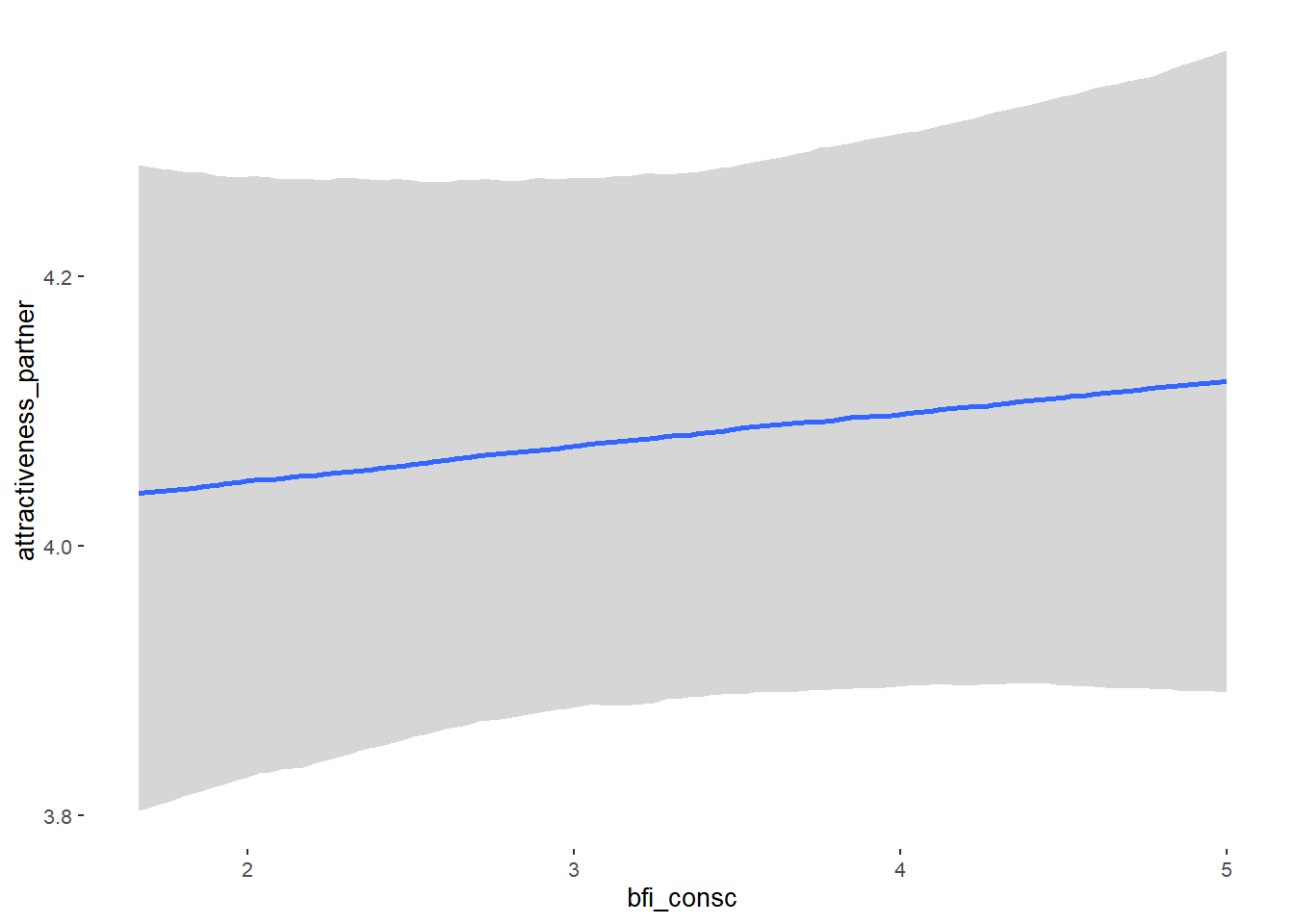
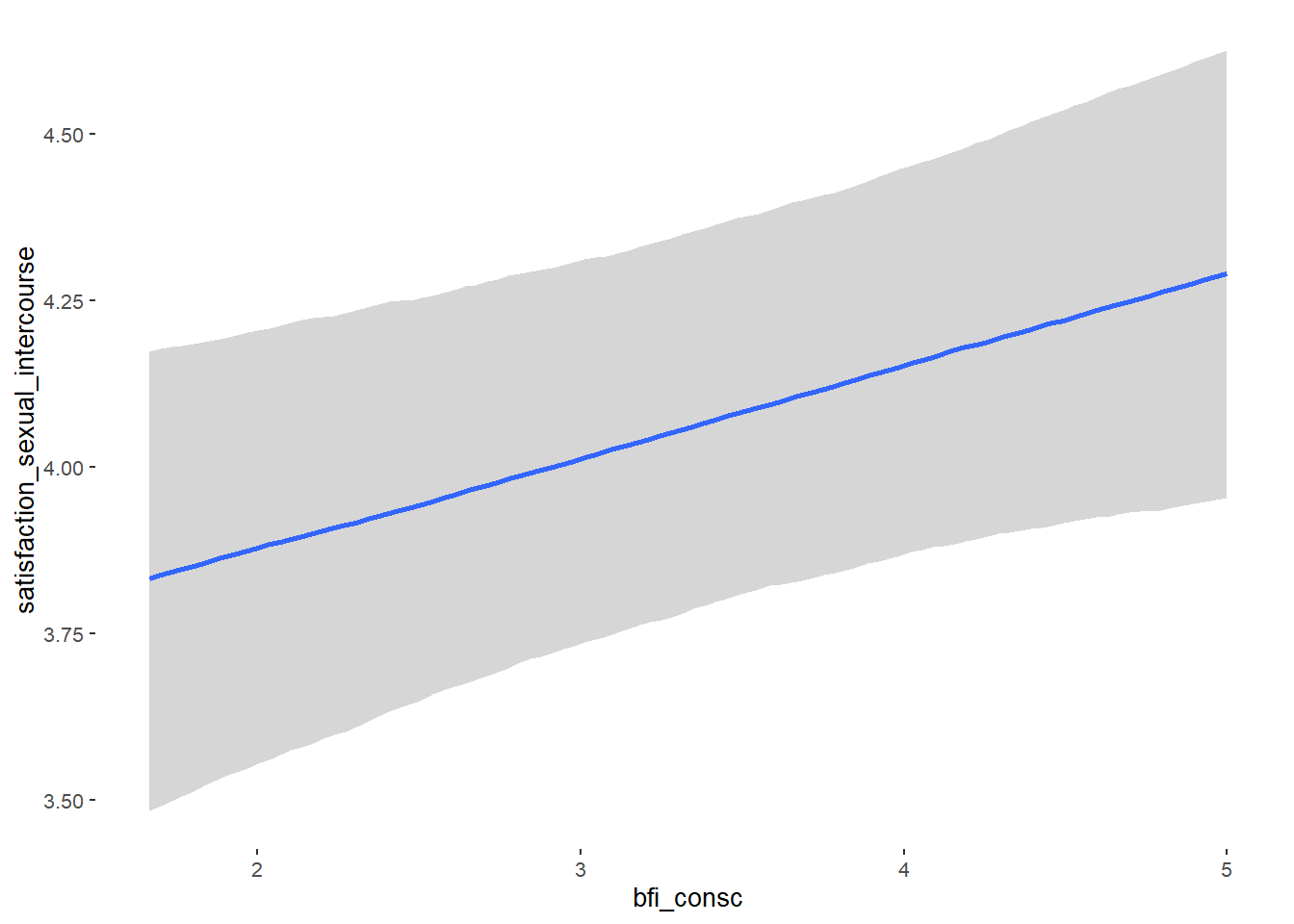
## bfi_consc 0.02 0.04 -0.05 0.09 1.09 40
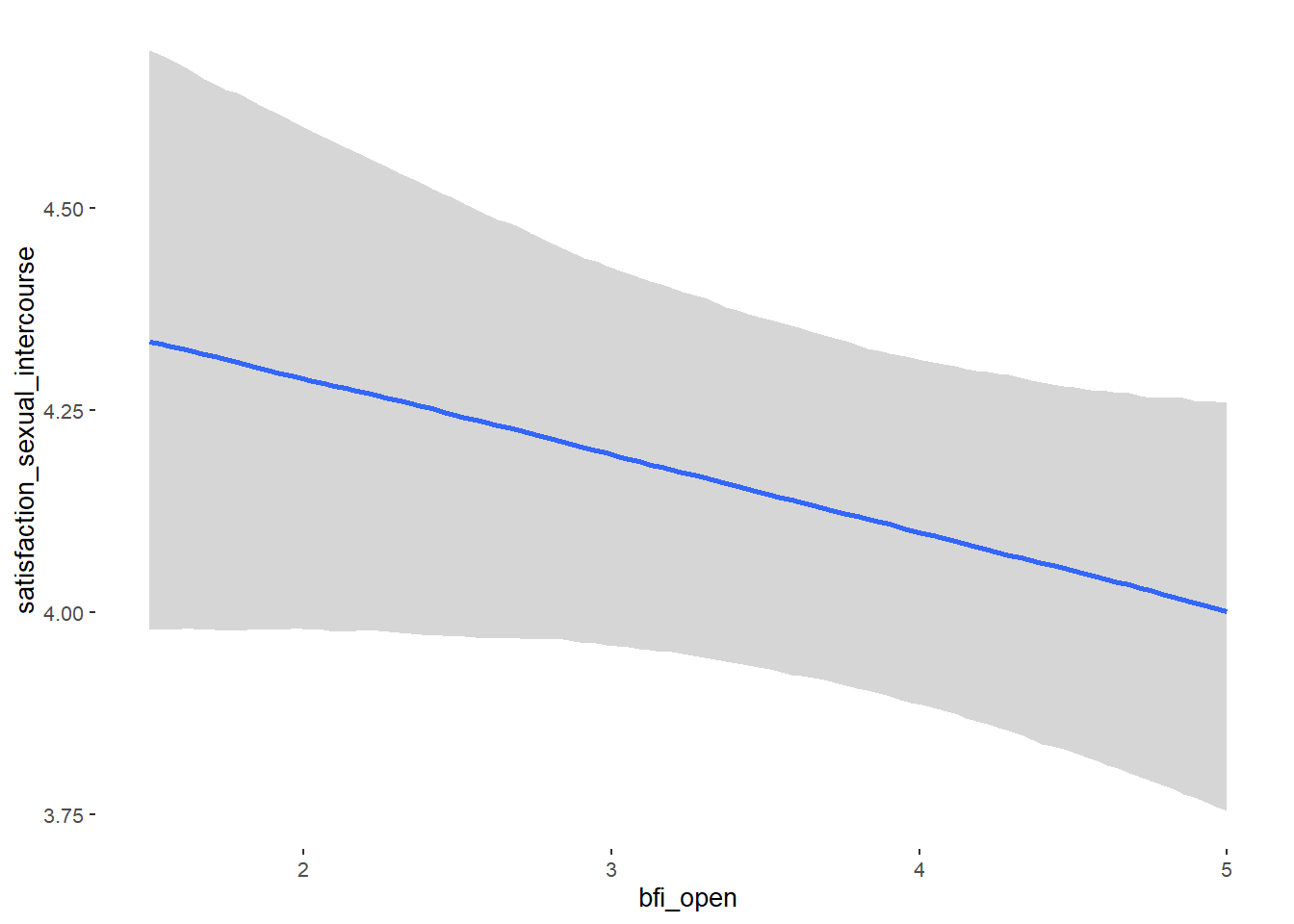
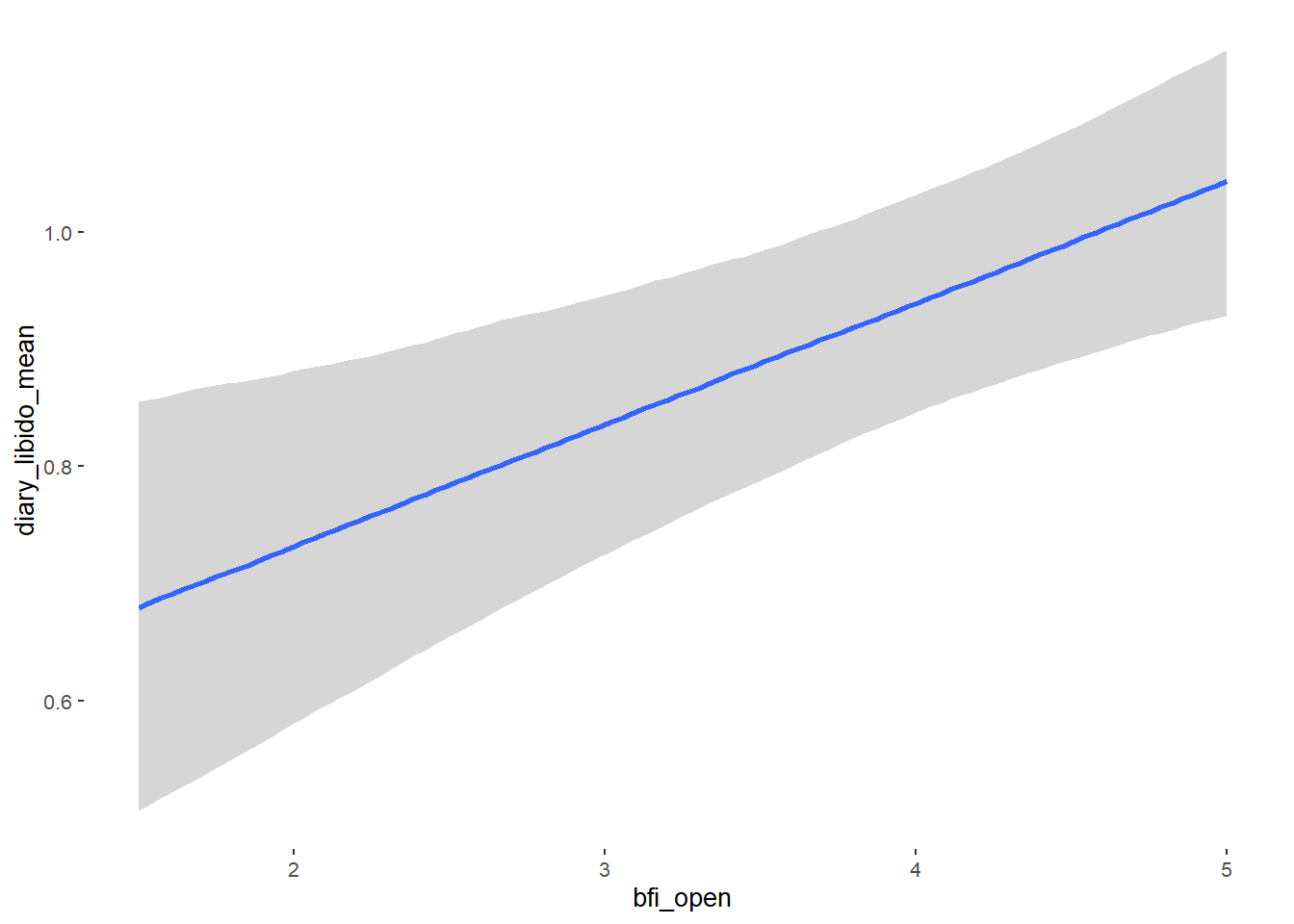
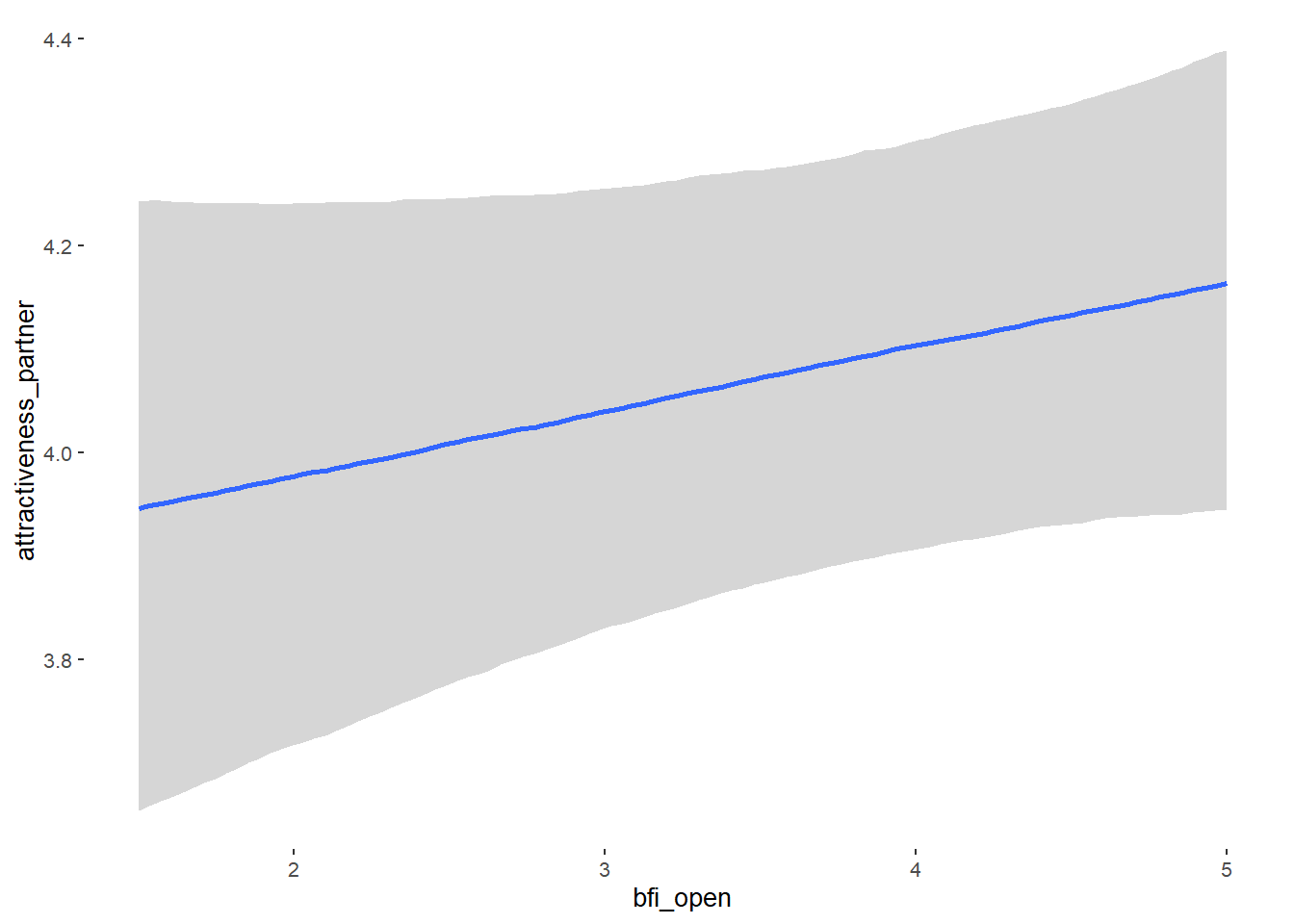
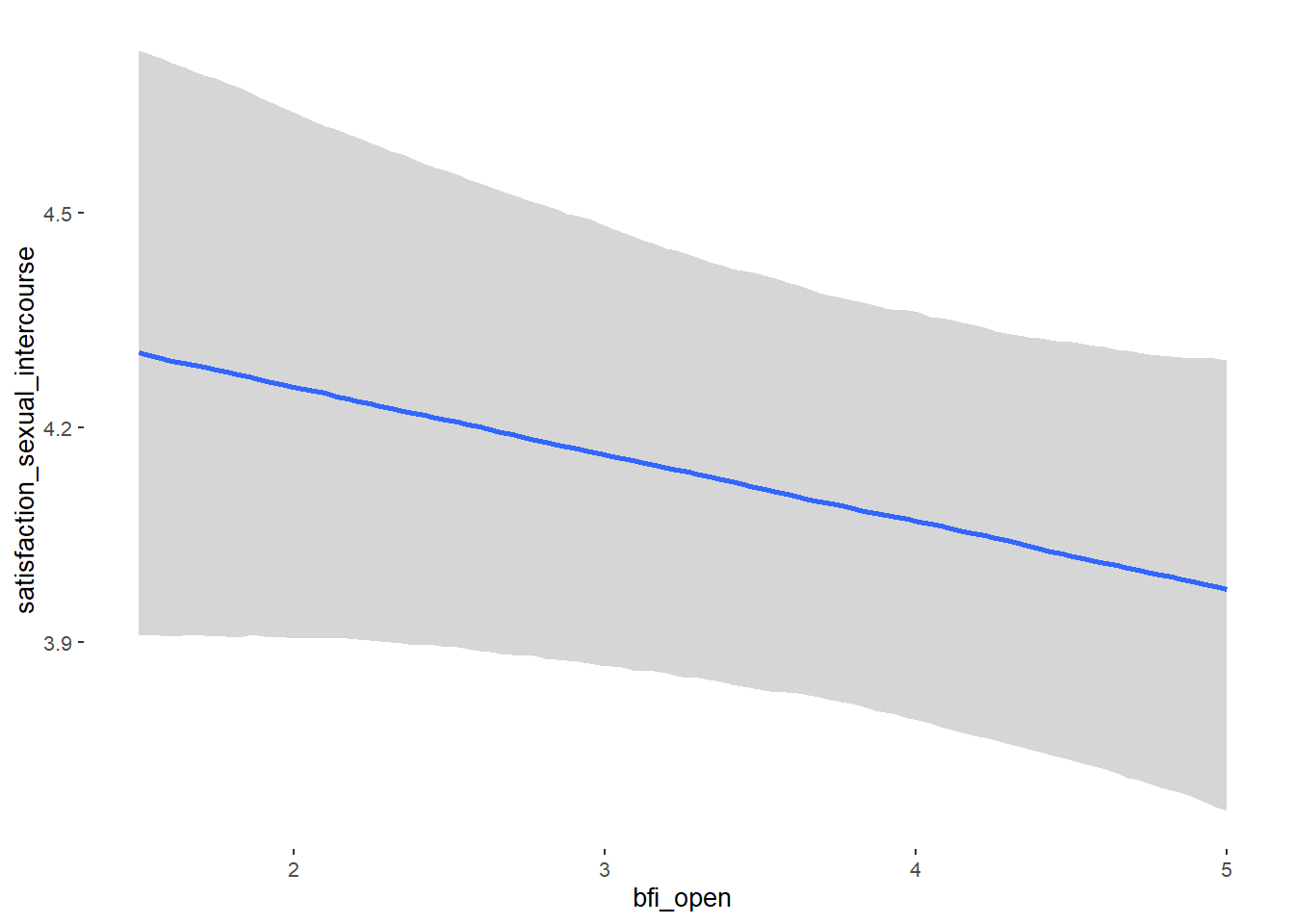
## bfi_open 0.07 0.04 0.01 0.14 1.09 53
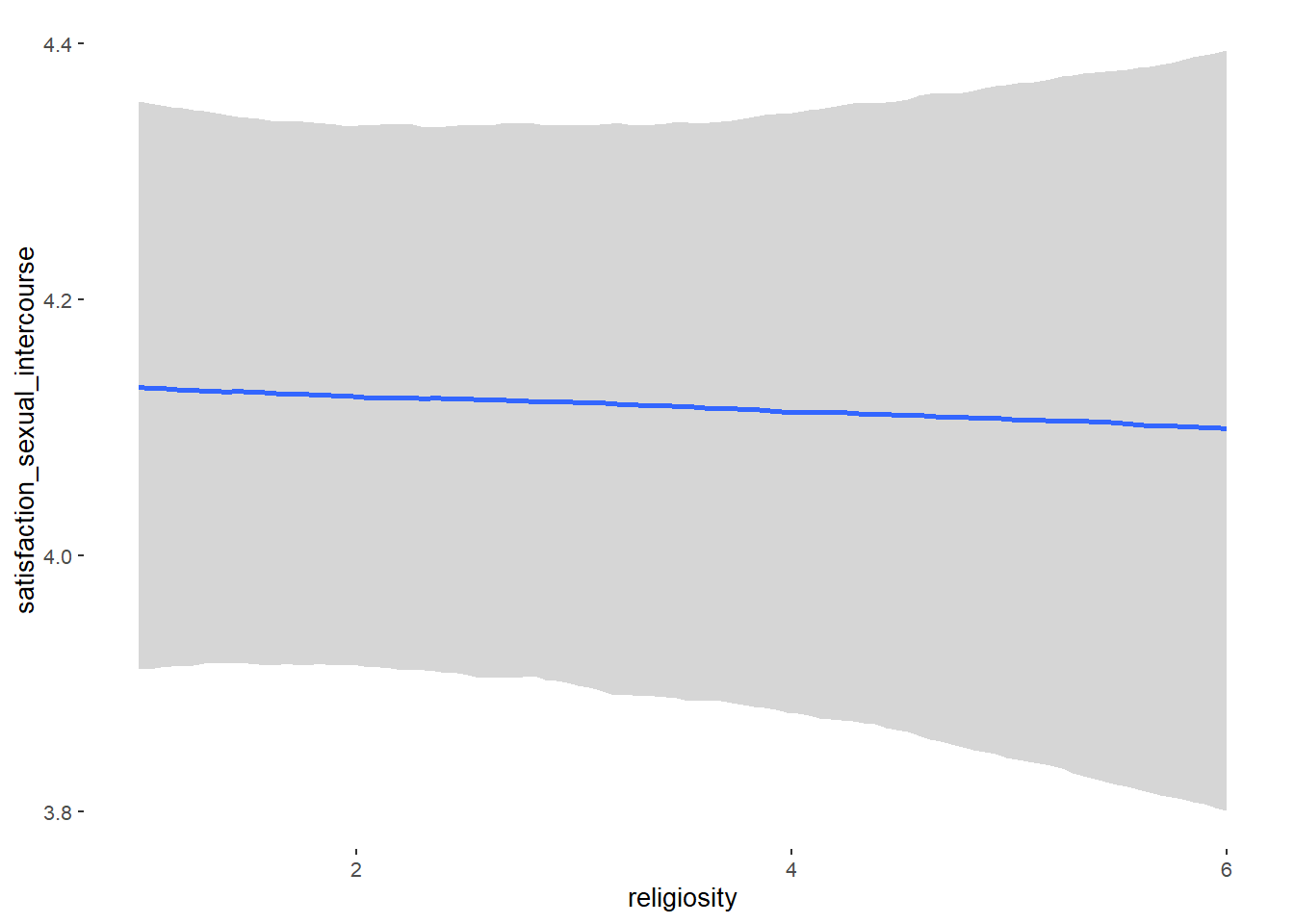
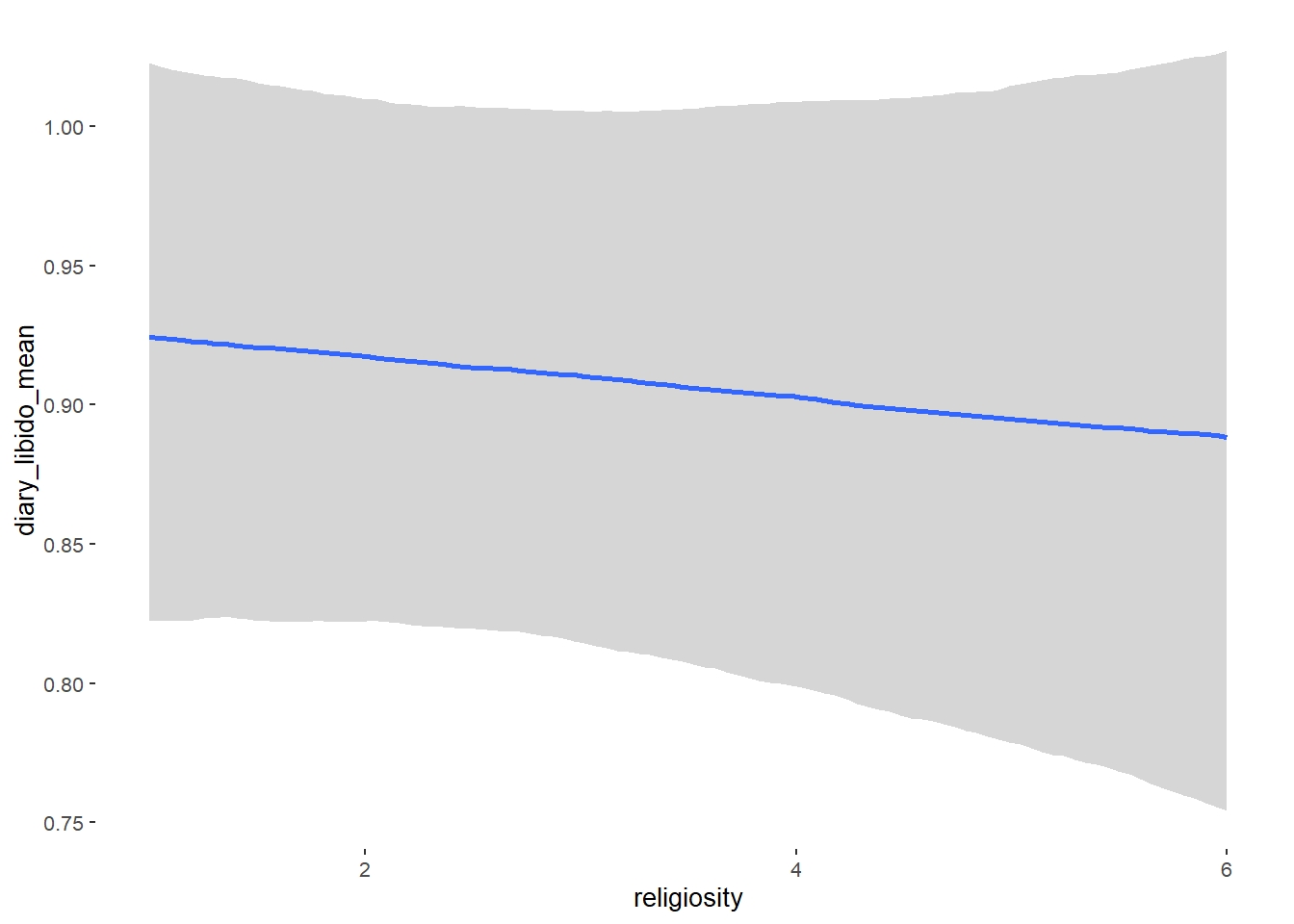
## religiosity -0.00 0.02 -0.03 0.03 1.03 52
## Tail_ESS
## Intercept NA
## contraception_hormonalyes NA
## age NA
## net_incomeeuro_500_1000 NA
## net_incomeeuro_1000_2000 NA
## net_incomeeuro_2000_3000 NA
## net_incomeeuro_gt_3000 NA
## net_incomedont_tell NA
## relationship_duration_factorPartnered_upto12months NA
## relationship_duration_factorPartnered_upto28months NA
## relationship_duration_factorPartnered_upto52months NA
## relationship_duration_factorPartnered_morethan52months NA
## education_years NA
## bfi_extra NA
## bfi_neuro NA
## bfi_agree NA
## bfi_consc NA
## bfi_open NA
## religiosity NA
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.73 0.02 0.69 0.76 1.22 15 NA
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_hc_atrr_controlled, range = c(-0.07, 0.07), ci = 0.90,
parameters = "contraception"))## Picking joint bandwidth of 0.00705## Warning: Removed 399 rows containing non-finite values (stat_density_ridges).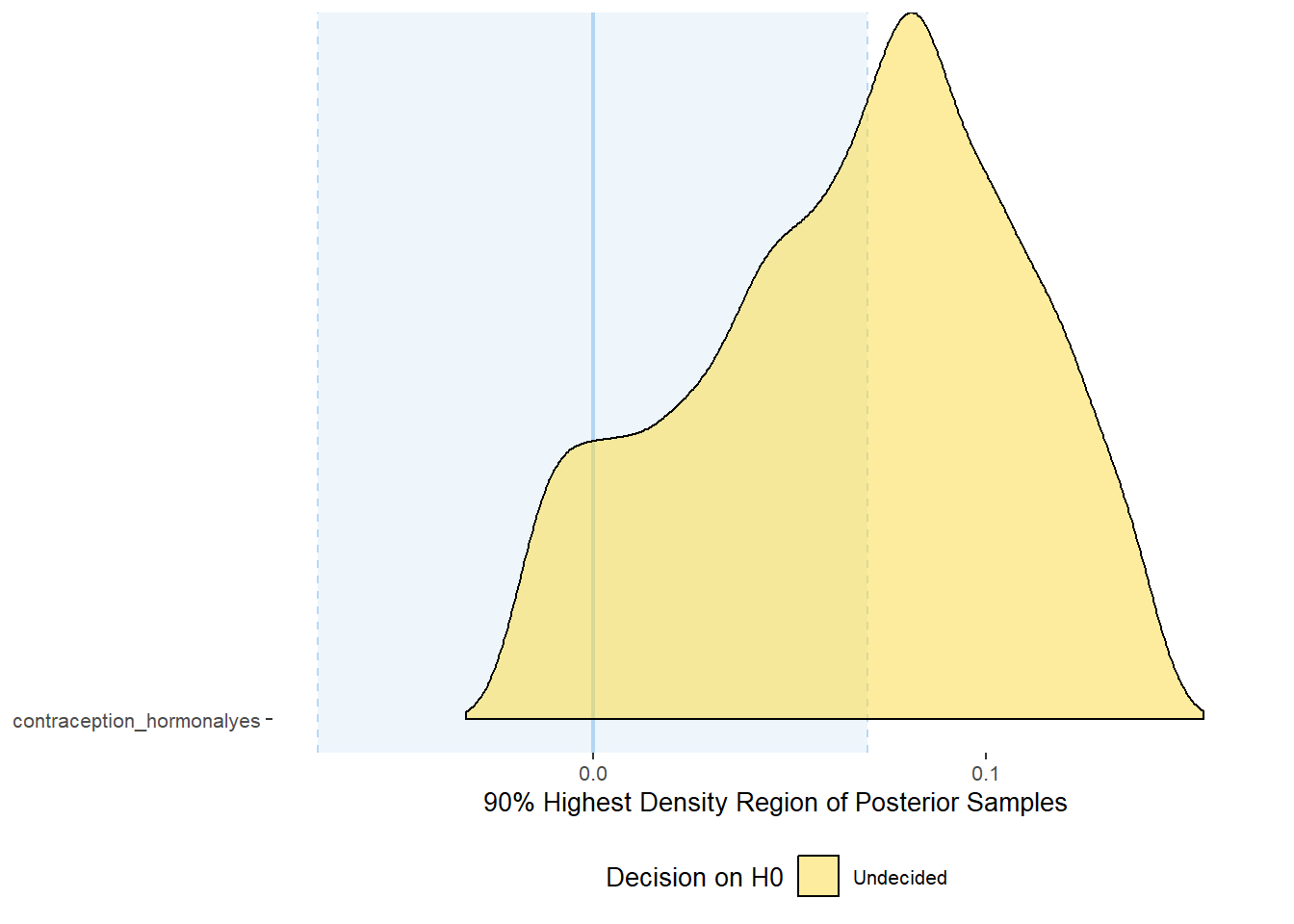
equivalence_test(m_hc_atrr_controlled, range = c(-0.07, 0.07), ci = 0.90,
parameters = "contraception")## # A tibble: 1 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.07 0.07 0.482 Undecided -0.0186 0.142 fixed conditio~Forest Plot for Effect Sizes
m_hc_atrr_controlled %>%
spread_draws(b_contraception_hormonalyes,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity) %>%
pivot_longer(cols = c(b_contraception_hormonalyes,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_relationship_duration_factor",
"Relationship Duration",
ifelse(condition %contains% "b_net_income",
"Income",
NA)),
group = ifelse(condition %contains% "b_contraception_hormonalyes",
"Contraception", group),
condition = ifelse(condition %contains% "b_contraception_hormonalyes",
"Hormonal Contraception", condition),
condition = ifelse(condition == "b_age", "Age",
ifelse(condition == "b_net_incomeeuro_500_1000", "500-1000 Euro",
ifelse(condition == "b_net_incomeeuro_1000_2000", "1000-2000 Euro",
ifelse(condition == "b_net_incomeeuro_2000_3000", "2000-3000 Euro",
ifelse(condition == "b_net_incomeeuro_gt_3000", ">3000 Euro",
ifelse(condition == "b_net_incomedont_tell", "do not tell",
ifelse(condition == "b_relationship_duration_factorPartnered_upto28months",
"13-28 months",
ifelse(condition == "b_relationship_duration_factorPartnered_upto52months",
"29-52 months",
ifelse(condition == "b_relationship_duration_factorPartnered_morethan52months",
">52 months",
ifelse(condition == "b_education_years", "Years of Education",
ifelse(condition == "b_bfi_extra", "Extraversion",
ifelse(condition == "b_bfi_neuro", "Neuroticism",
ifelse(condition == "b_bfi_agree", "Agreeableness",
ifelse(condition == "b_bfi_consc", "Conscientiousness",
ifelse(condition == "b_bfi_open", "Openness",
ifelse(condition == "b_religiosity", "Religiosity",
condition)))))))))))))))),
group = ifelse(is.na(group), condition, group),
condition = factor(condition, levels = rev(c("Hormonal Contraception", "Age",
"500-1000 Euro", "1000-2000 Euro",
"2000-3000 Euro", ">3000 Euro", "do not tell",
"13-28 months", "29-52 months",
">52 months",
"Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))),
group = factor(group, levels = c("Contraception", "Age", "Income",
"Relationship Duration","Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.07))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.07, 0.07), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")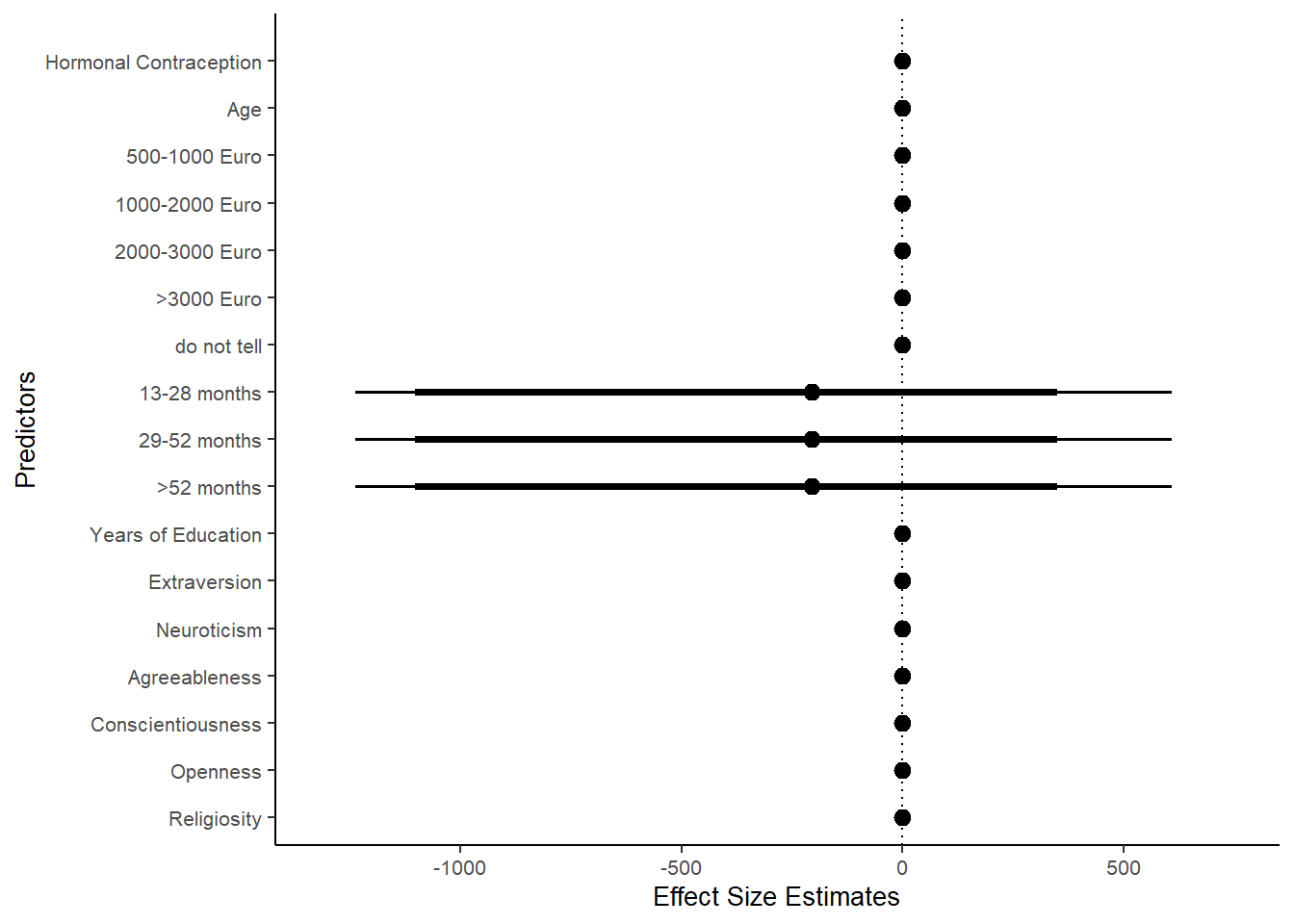
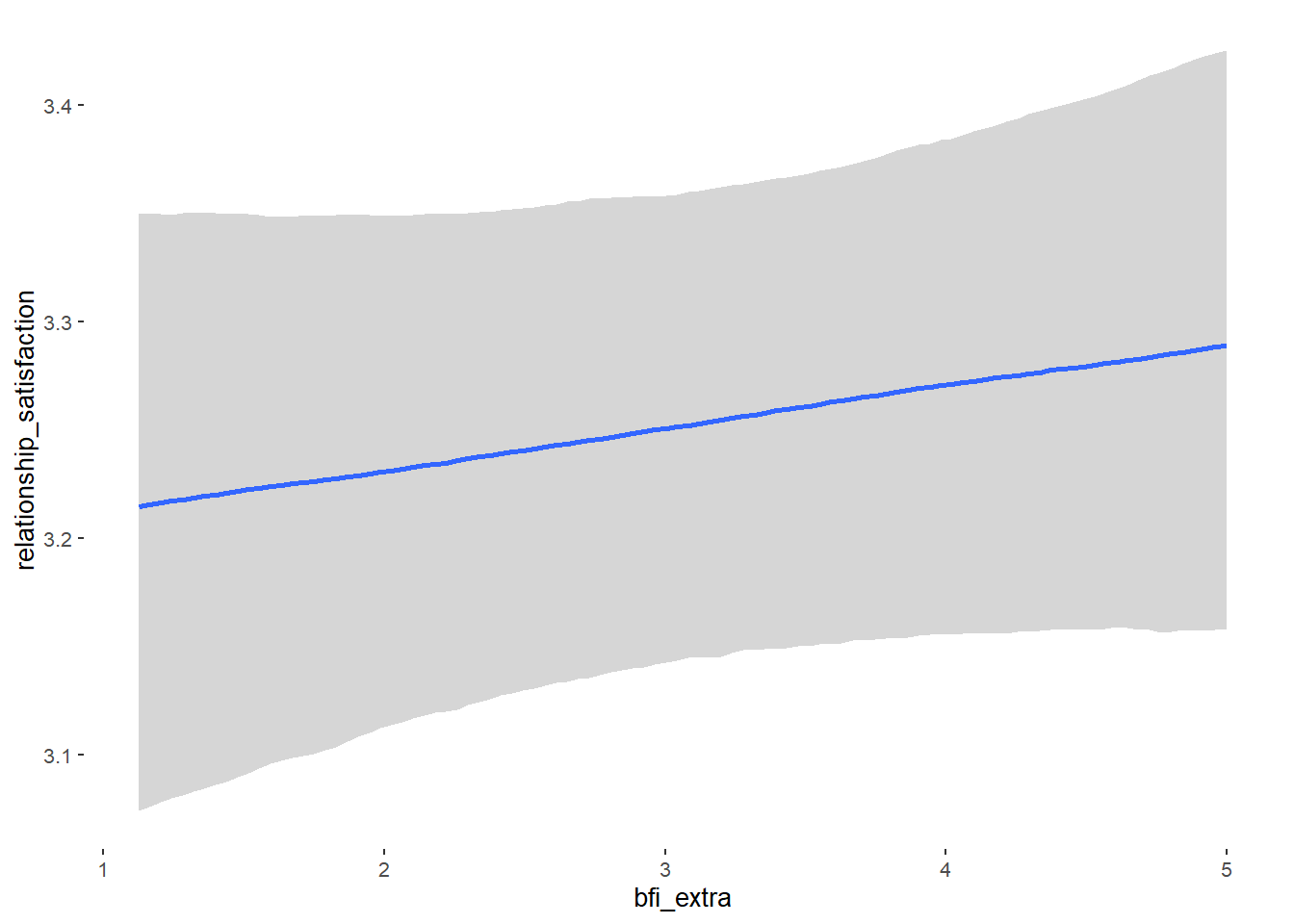
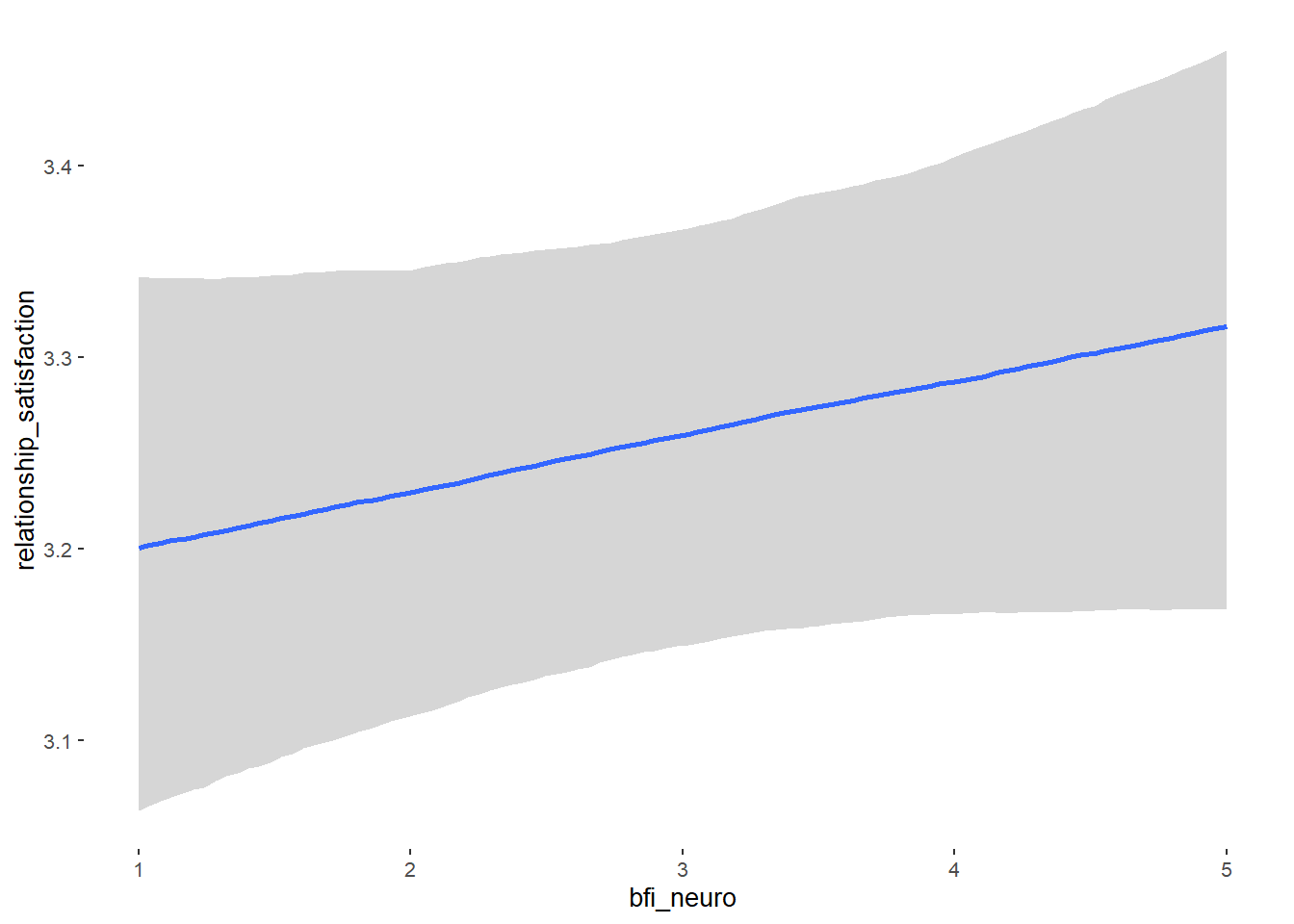
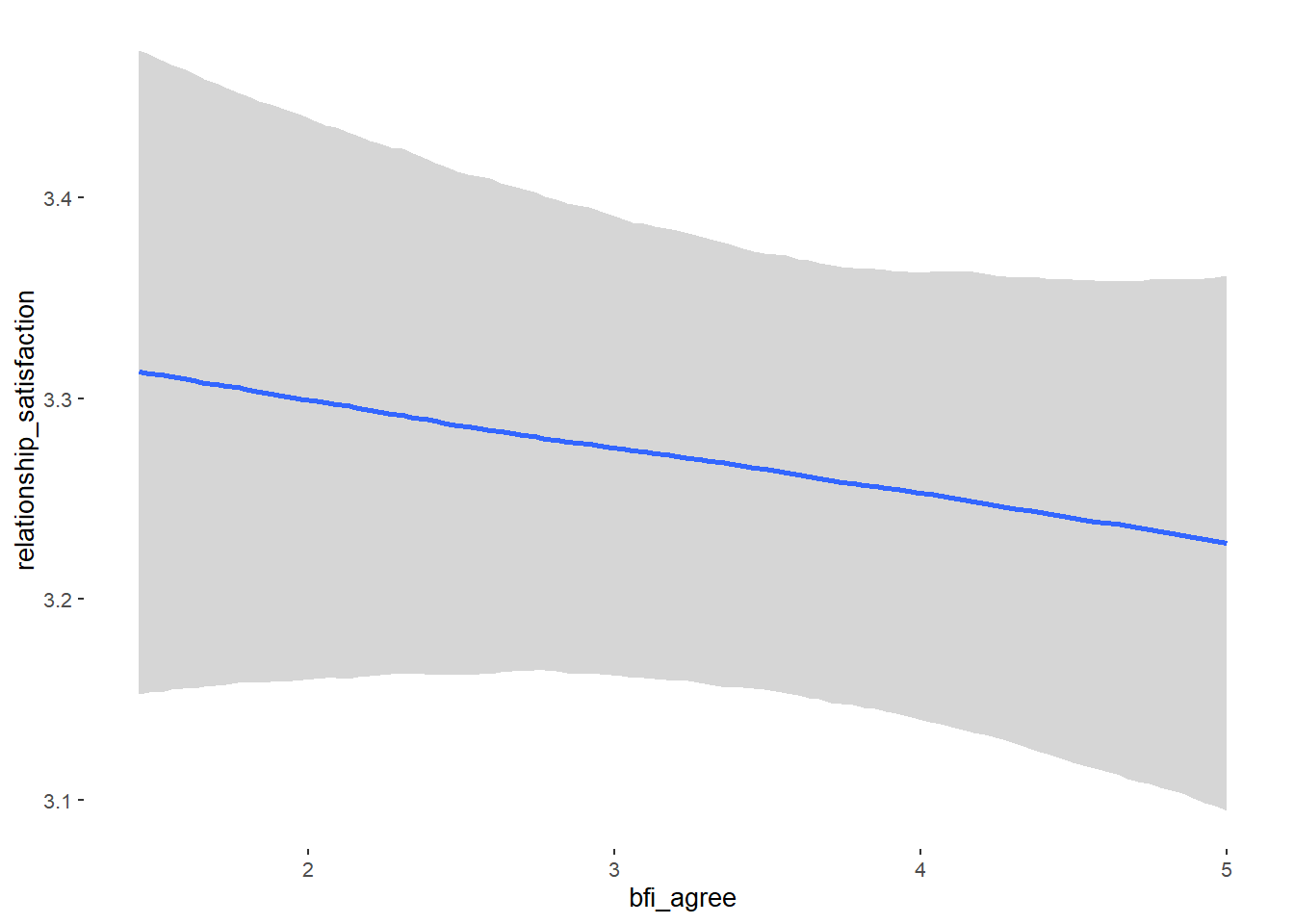
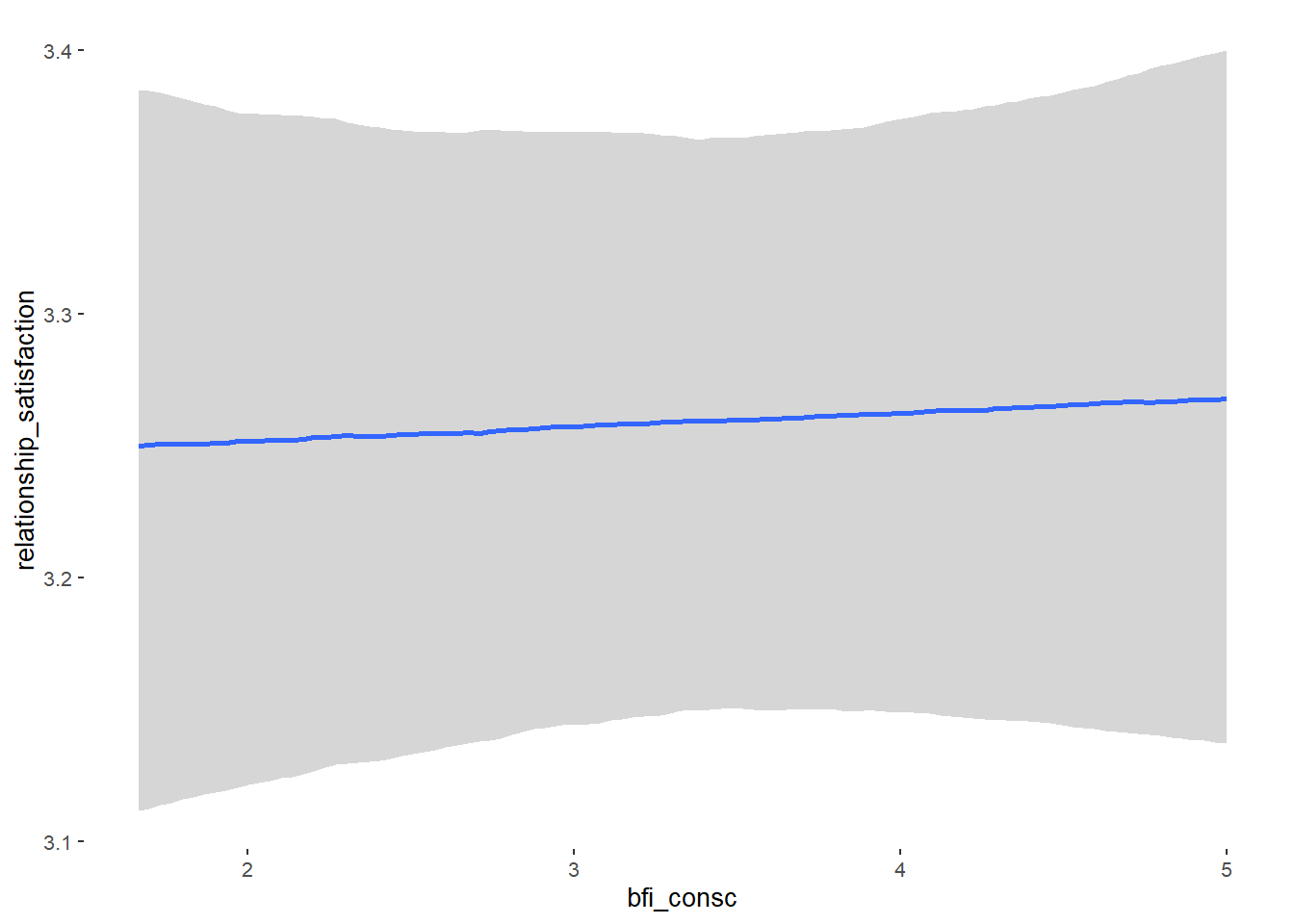
Relationship Satisfaction
Model
Summary
## Family: gaussian
## Links: mu = identity; sigma = identity
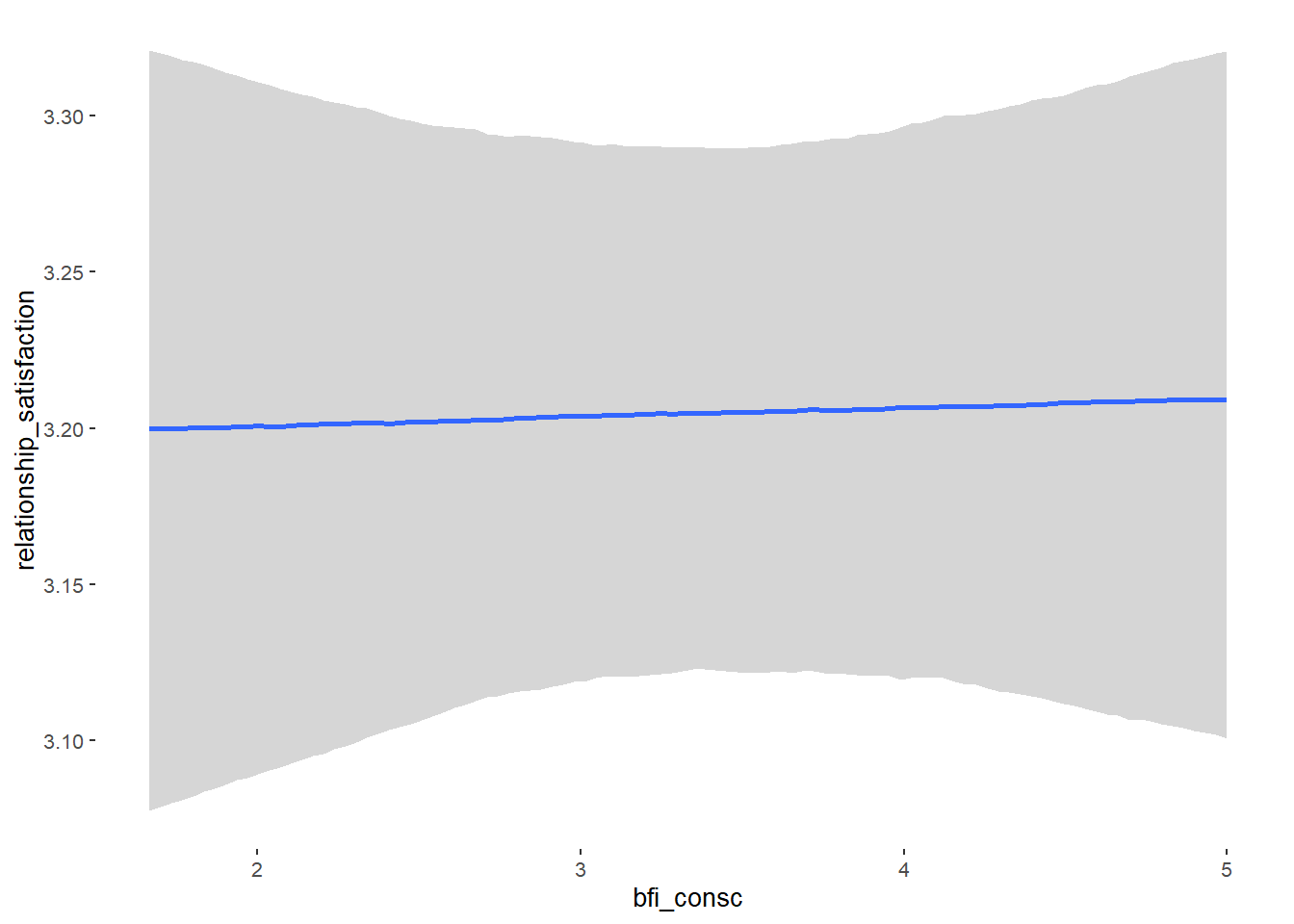
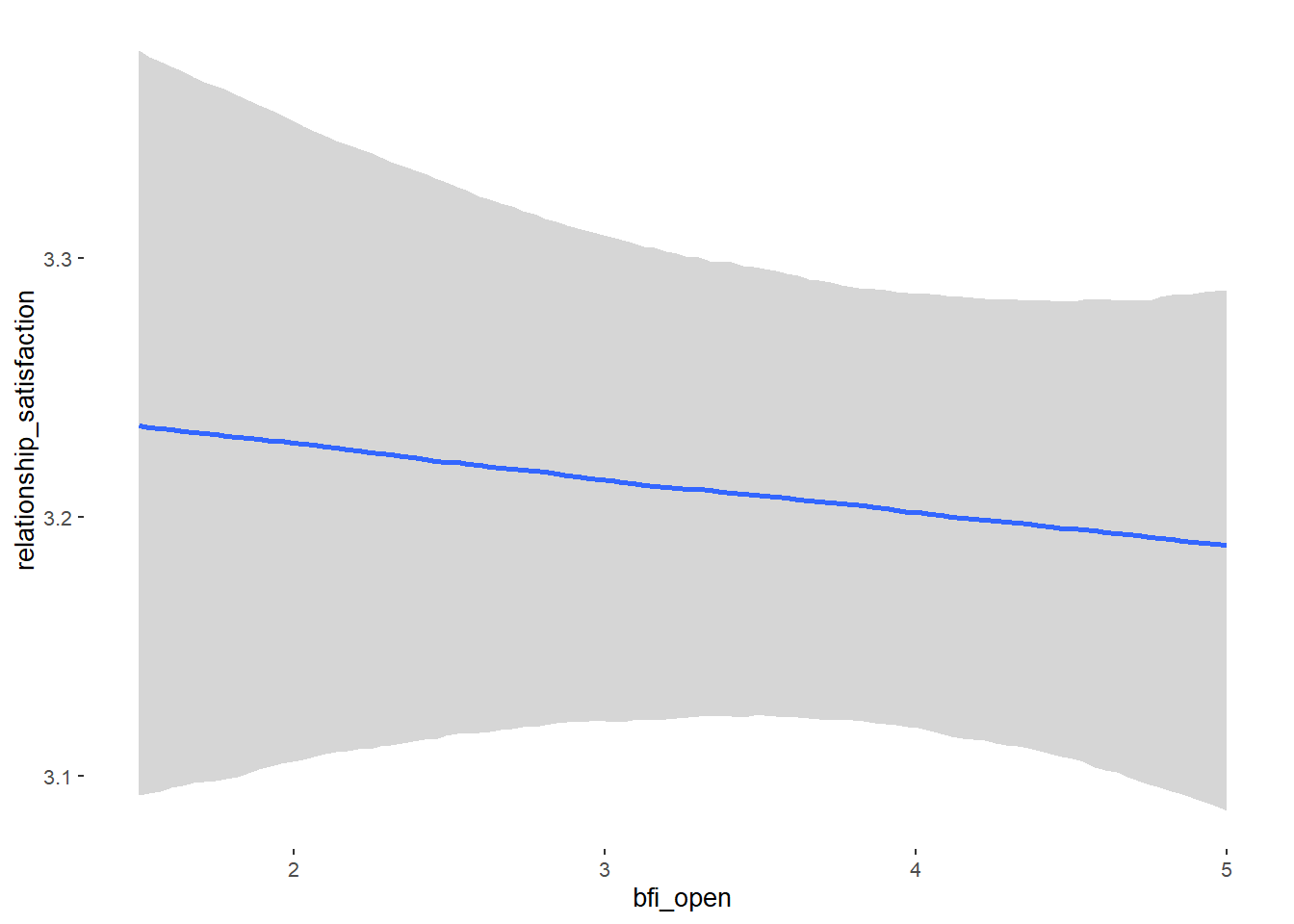
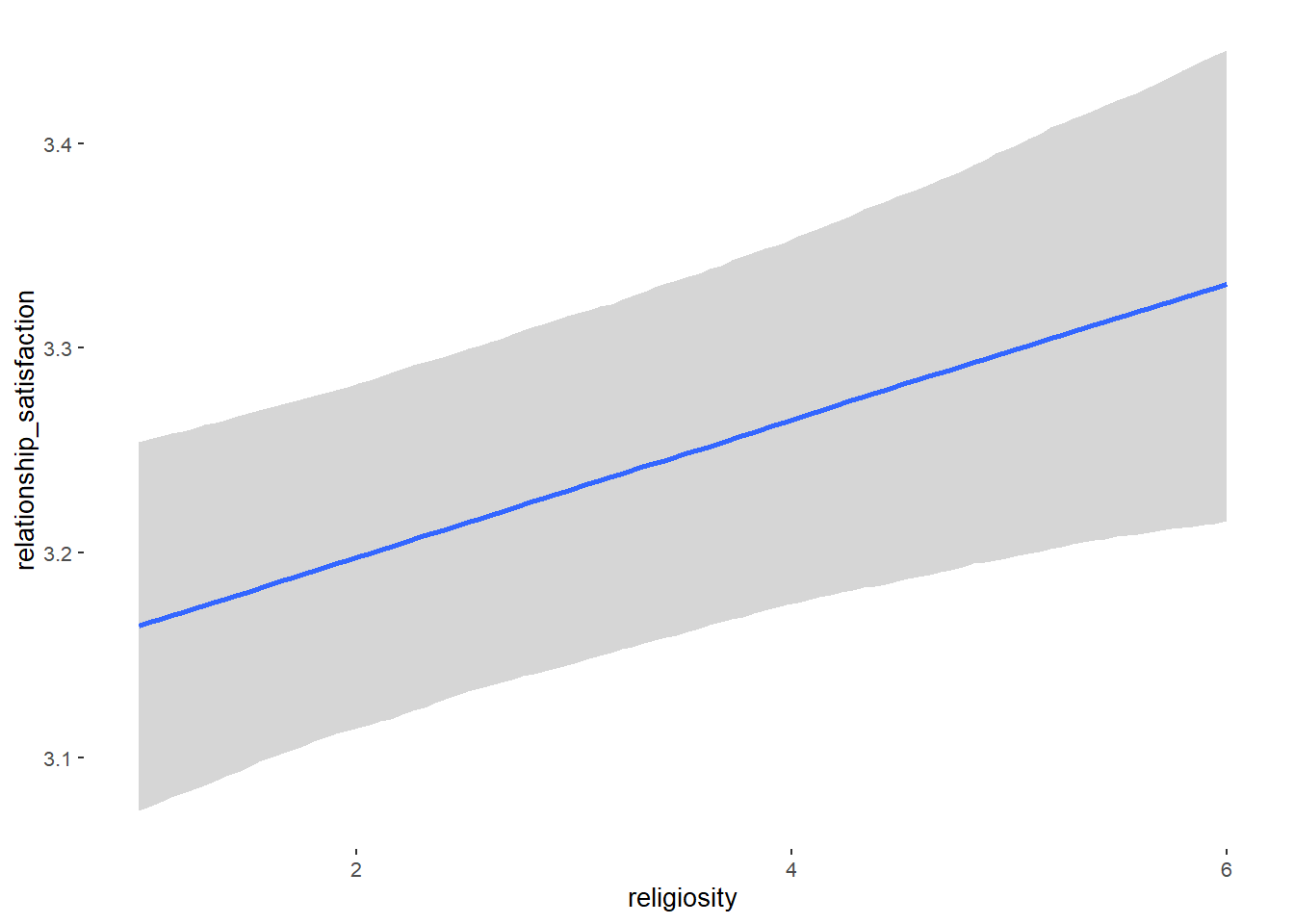
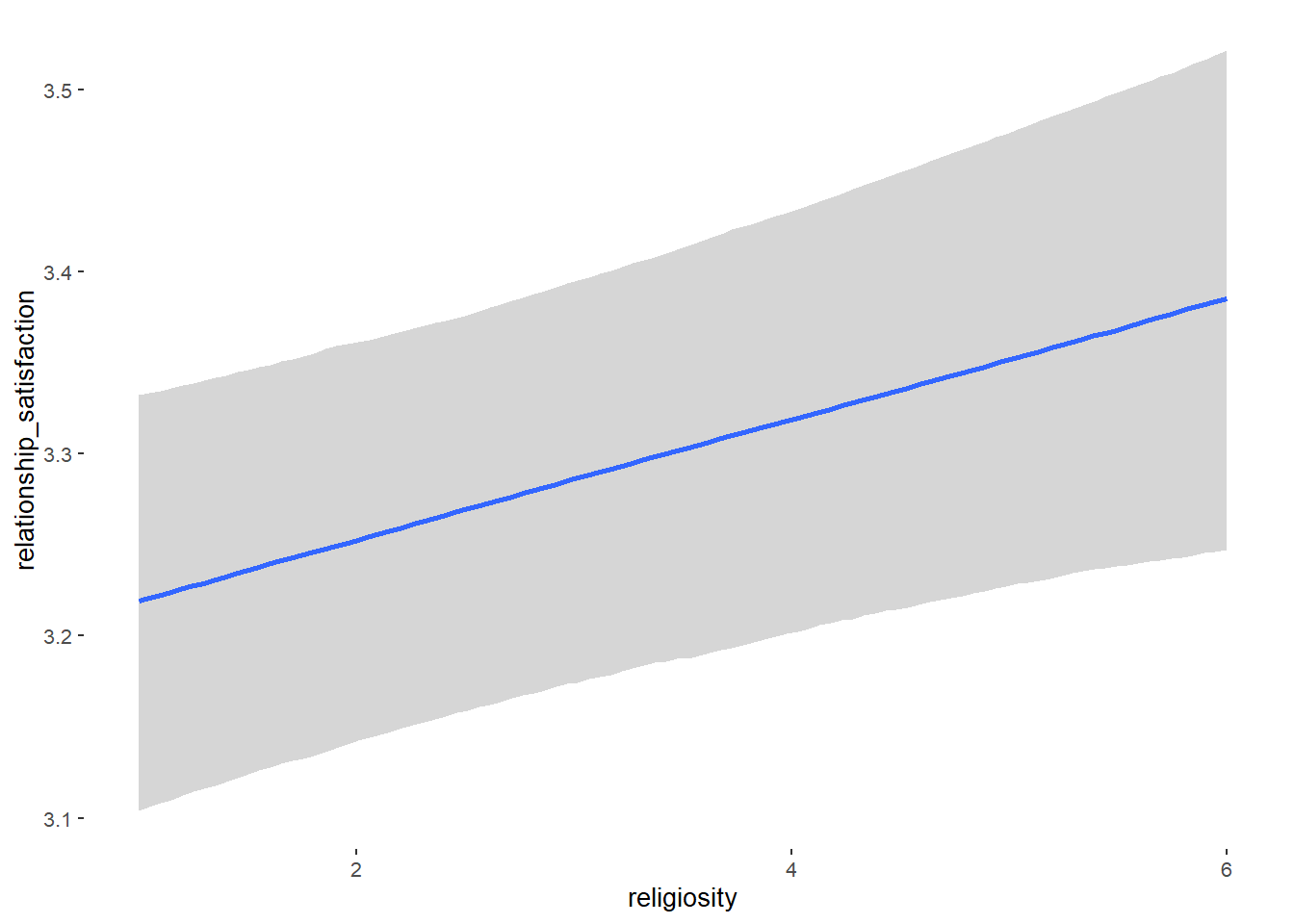
## Formula: relationship_satisfaction ~ contraception_hormonal + age + net_income + relationship_duration_factor + education_years + bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open + religiosity
## Data: data (Number of observations: 774)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept 3.29 0.22 2.92 3.64 1.00 4495
## contraception_hormonalyes 0.06 0.03 0.00 0.11 1.00 4877
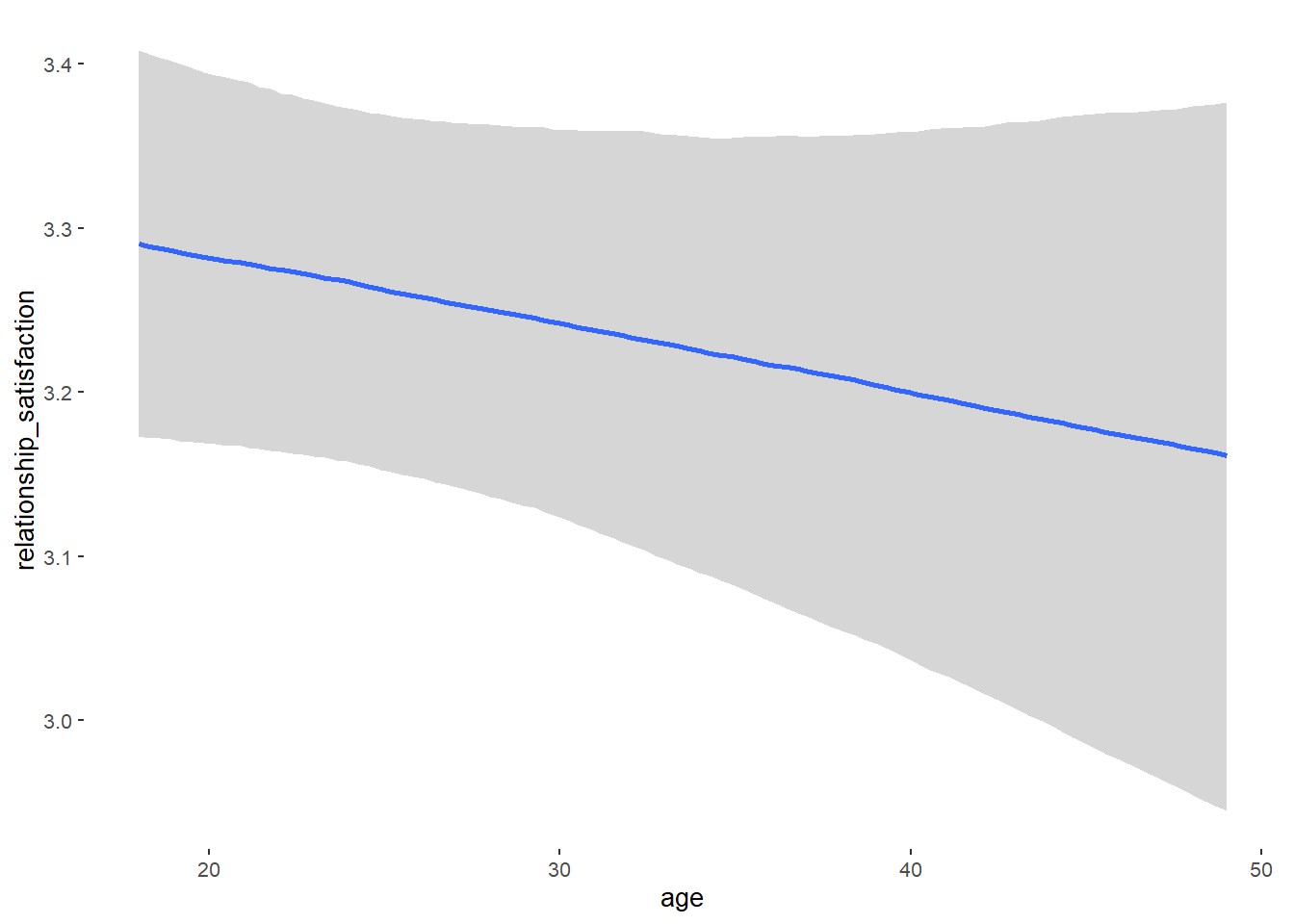
## age -0.00 0.00 -0.01 0.00 1.00 4011
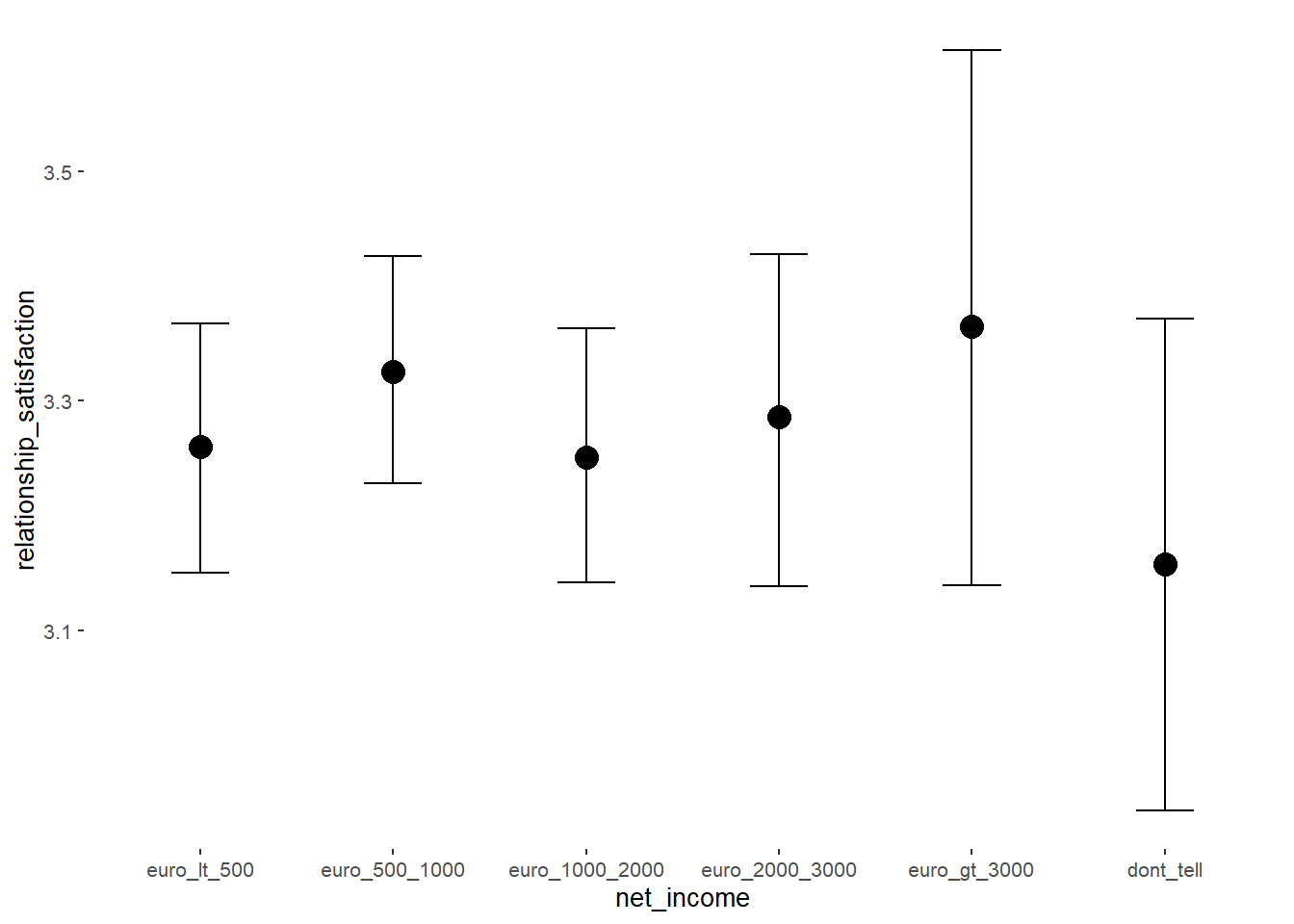
## net_incomeeuro_500_1000 0.07 0.04 0.01 0.13 1.00 3190
## net_incomeeuro_1000_2000 -0.01 0.05 -0.09 0.08 1.00 2692
## net_incomeeuro_2000_3000 0.04 0.07 -0.08 0.15 1.00 3233
## net_incomeeuro_gt_3000 0.11 0.12 -0.08 0.31 1.00 3668
## net_incomedont_tell -0.10 0.10 -0.26 0.07 1.00 4044
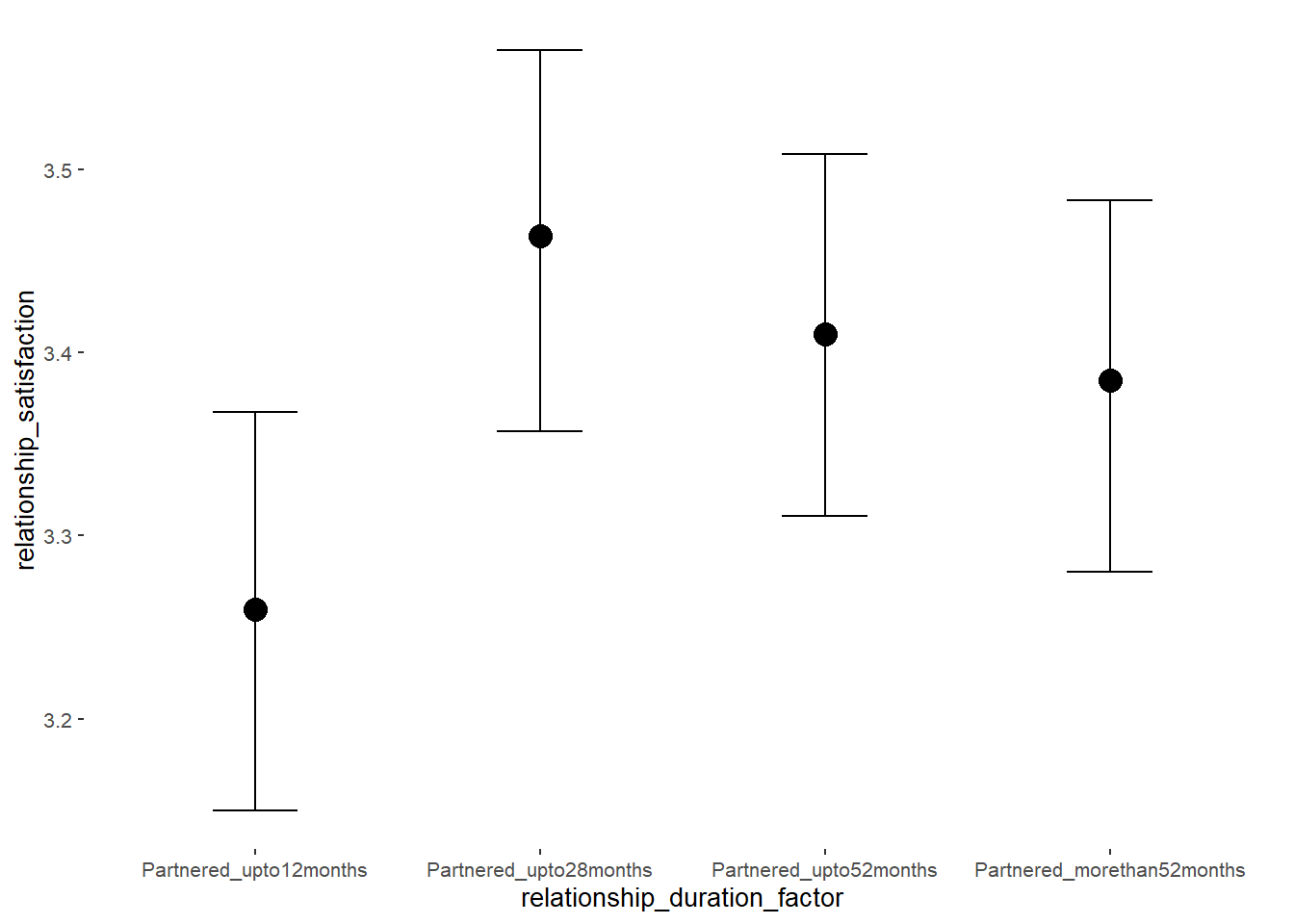
## relationship_duration_factorPartnered_upto28months 0.21 0.04 0.14 0.28 1.00 3251
## relationship_duration_factorPartnered_upto52months 0.17 0.04 0.10 0.24 1.00 3109
## relationship_duration_factorPartnered_morethan52months 0.14 0.04 0.07 0.21 1.00 3319
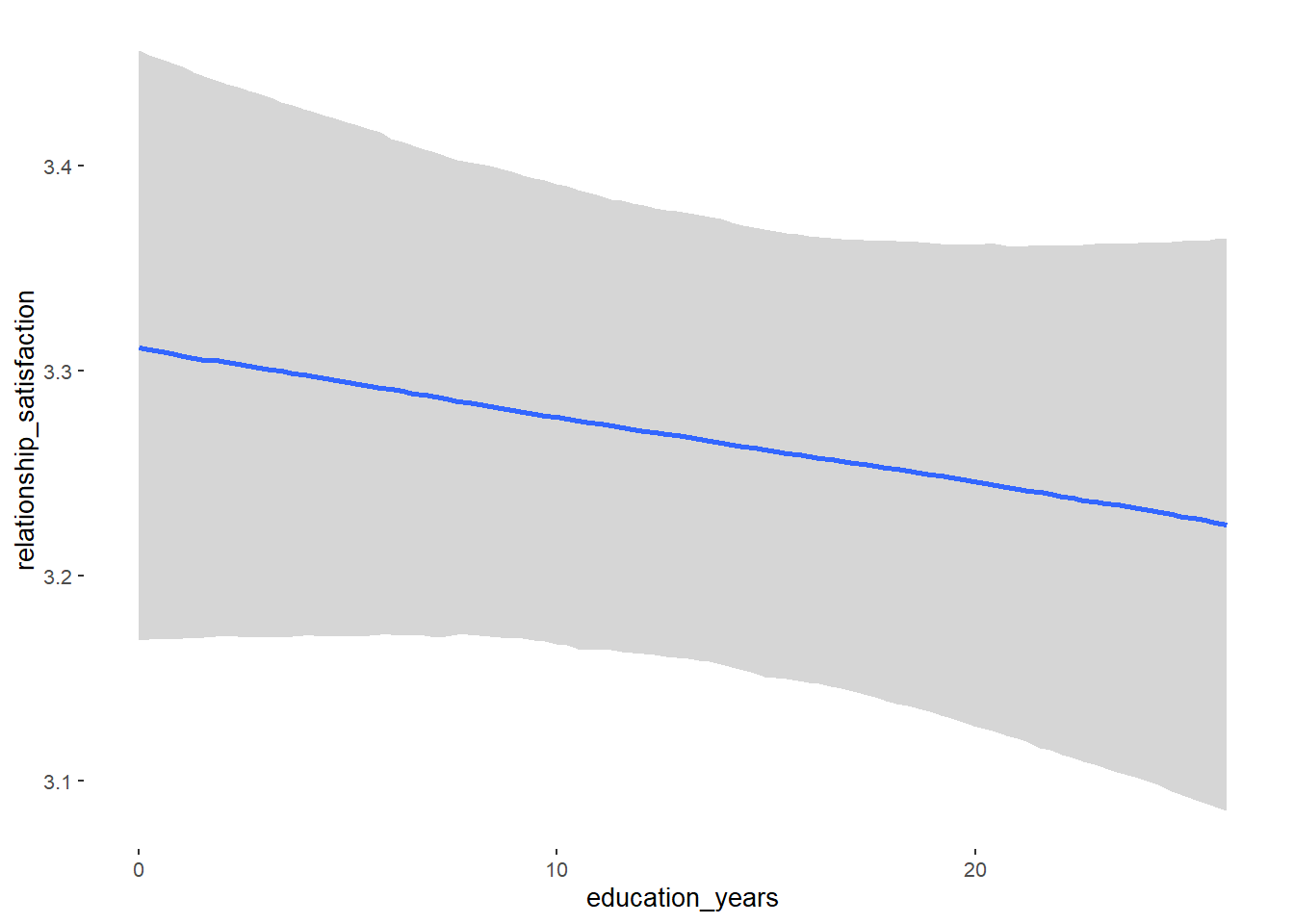
## education_years -0.00 0.00 -0.01 0.00 1.00 7198
## bfi_extra 0.02 0.02 -0.01 0.05 1.00 4801
## bfi_neuro 0.03 0.02 -0.01 0.07 1.00 4013
## bfi_agree -0.02 0.03 -0.07 0.02 1.00 3944
## bfi_consc 0.00 0.02 -0.04 0.04 1.00 4361
## bfi_open -0.01 0.03 -0.06 0.03 1.00 4640
## religiosity 0.03 0.01 0.02 0.05 1.00 5147
## Tail_ESS
## Intercept 3130
## contraception_hormonalyes 3101
## age 3585
## net_incomeeuro_500_1000 3354
## net_incomeeuro_1000_2000 3107
## net_incomeeuro_2000_3000 3115
## net_incomeeuro_gt_3000 3275
## net_incomedont_tell 3320
## relationship_duration_factorPartnered_upto28months 3199
## relationship_duration_factorPartnered_upto52months 3115
## relationship_duration_factorPartnered_morethan52months 3246
## education_years 2915
## bfi_extra 3333
## bfi_neuro 2932
## bfi_agree 2854
## bfi_consc 2822
## bfi_open 3049
## religiosity 3279
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.41 0.01 0.40 0.43 1.00 5262 3051
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_hc_relsat_controlled, range = c(-0.04, 0.04), ci = 0.90,
parameters = "contraception"))## Picking joint bandwidth of 0.00437## Warning: Removed 399 rows containing non-finite values (stat_density_ridges).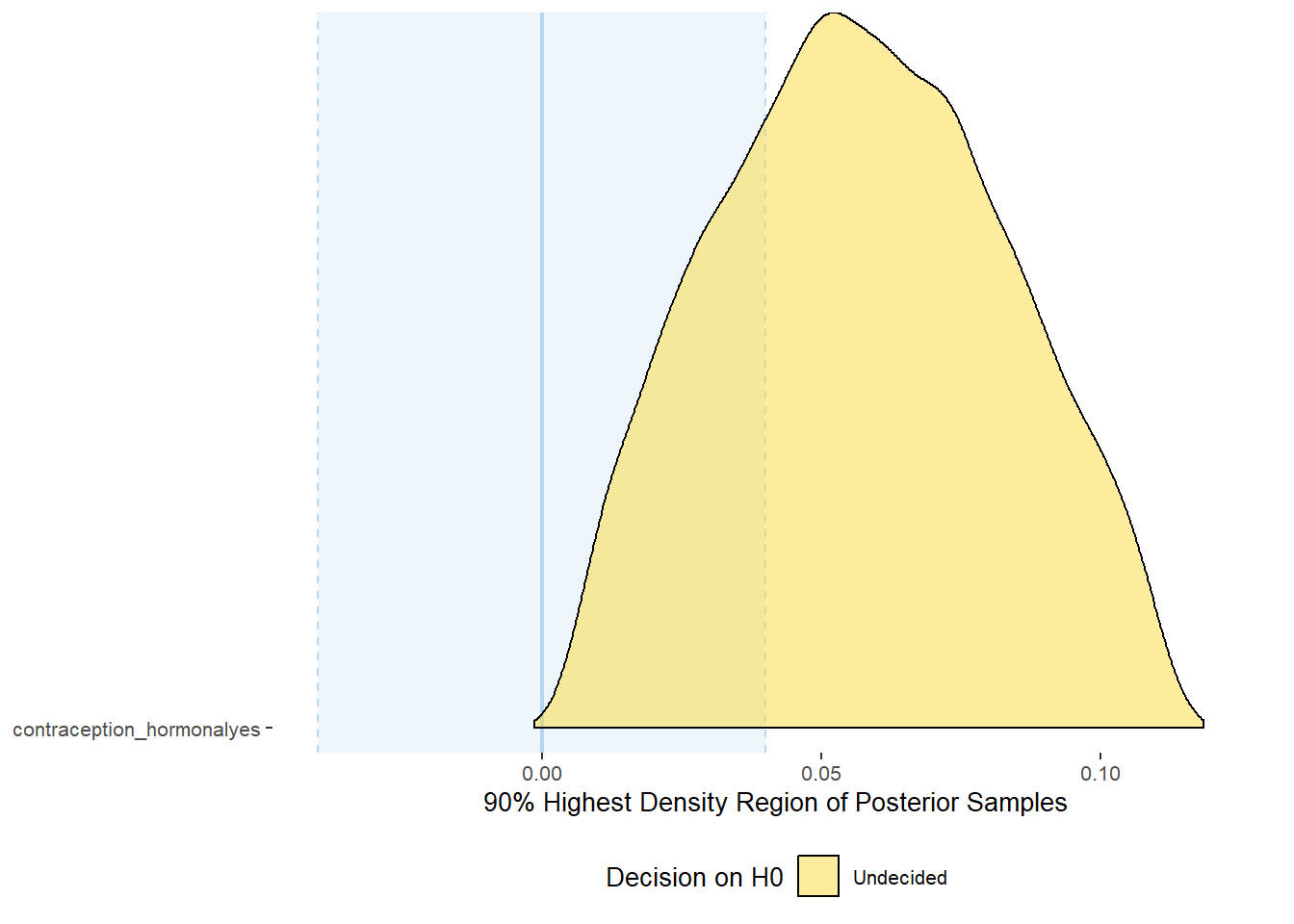
equivalence_test(m_hc_relsat_controlled, range = c(-0.04, 0.04), ci = 0.90,
parameters = "contraception")## # A tibble: 1 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.04 0.04 0.275 Undecided 0.00646 0.111 fixed conditio~Forest Plot for Effect Sizes
m_hc_relsat_controlled %>%
spread_draws(b_contraception_hormonalyes,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity) %>%
pivot_longer(cols = c(b_contraception_hormonalyes,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_relationship_duration_factor",
"Relationship Duration",
ifelse(condition %contains% "b_net_income",
"Income",
NA)),
group = ifelse(condition %contains% "b_contraception_hormonalyes",
"Contraception", group),
condition = ifelse(condition %contains% "b_contraception_hormonalyes",
"Hormonal Contraception", condition),
condition = ifelse(condition == "b_age", "Age",
ifelse(condition == "b_net_incomeeuro_500_1000", "500-1000 Euro",
ifelse(condition == "b_net_incomeeuro_1000_2000", "1000-2000 Euro",
ifelse(condition == "b_net_incomeeuro_2000_3000", "2000-3000 Euro",
ifelse(condition == "b_net_incomeeuro_gt_3000", ">3000 Euro",
ifelse(condition == "b_net_incomedont_tell", "do not tell",
ifelse(condition == "b_relationship_duration_factorPartnered_upto28months",
"13-28 months",
ifelse(condition == "b_relationship_duration_factorPartnered_upto52months",
"29-52 months",
ifelse(condition == "b_relationship_duration_factorPartnered_morethan52months",
">52 months",
ifelse(condition == "b_education_years", "Years of Education",
ifelse(condition == "b_bfi_extra", "Extraversion",
ifelse(condition == "b_bfi_neuro", "Neuroticism",
ifelse(condition == "b_bfi_agree", "Agreeableness",
ifelse(condition == "b_bfi_consc", "Conscientiousness",
ifelse(condition == "b_bfi_open", "Openness",
ifelse(condition == "b_religiosity", "Religiosity",
condition)))))))))))))))),
group = ifelse(is.na(group), condition, group),
condition = factor(condition, levels = rev(c("Hormonal Contraception", "Age",
"500-1000 Euro", "1000-2000 Euro",
"2000-3000 Euro", ">3000 Euro", "do not tell",
"13-28 months", "29-52 months",
">52 months",
"Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))),
group = factor(group, levels = c("Contraception", "Age", "Income",
"Relationship Duration","Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.04))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.04, 0.04), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")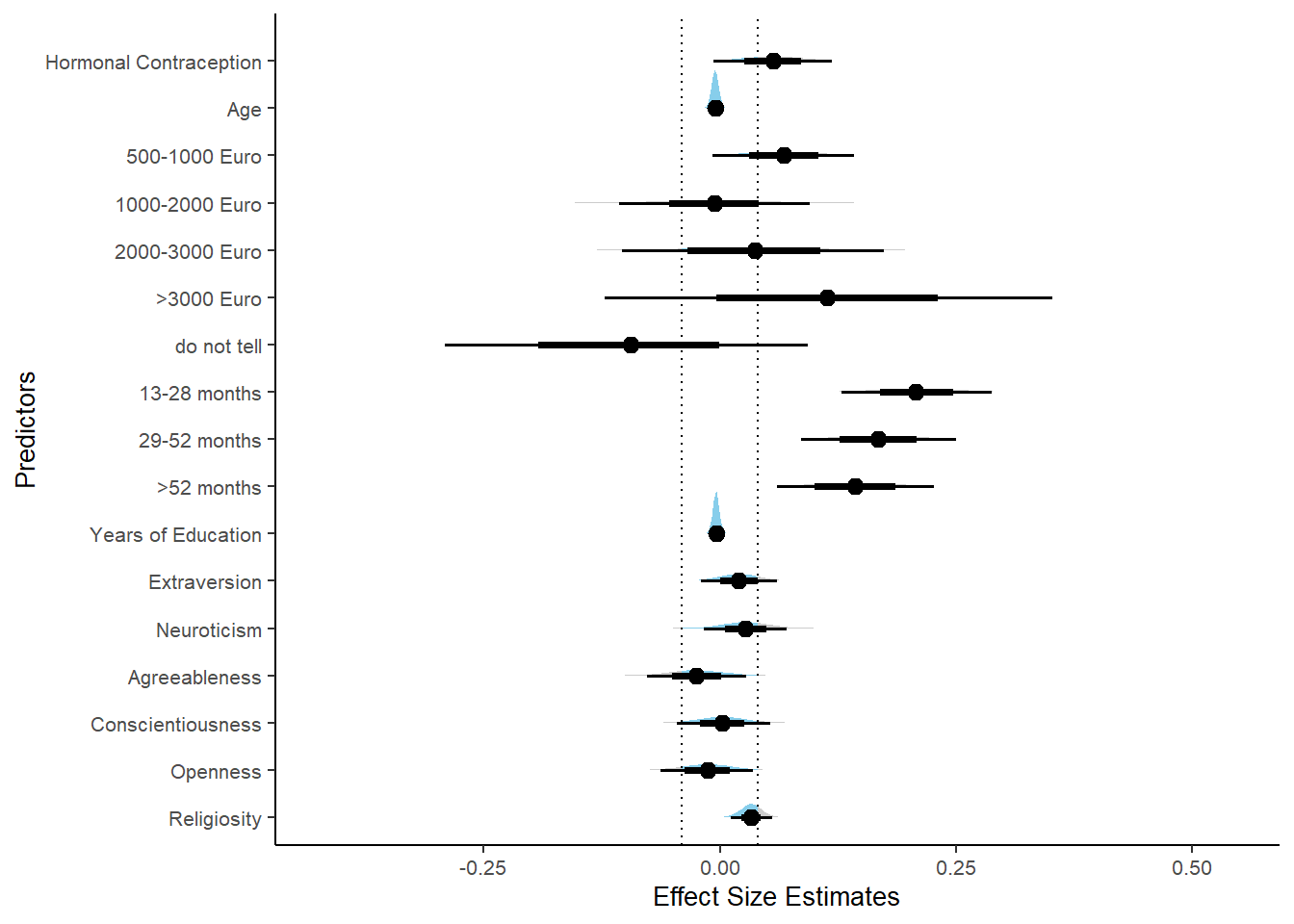
Sexual Satisfaction
Model
Summary
## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: satisfaction_sexual_intercourse ~ contraception_hormonal + age + net_income + relationship_duration_factor + education_years + bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open + religiosity
## Data: data (Number of observations: 774)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept 3.20 0.53 2.35 4.05 1.00 4370
## contraception_hormonalyes 0.11 0.08 -0.02 0.24 1.00 4602
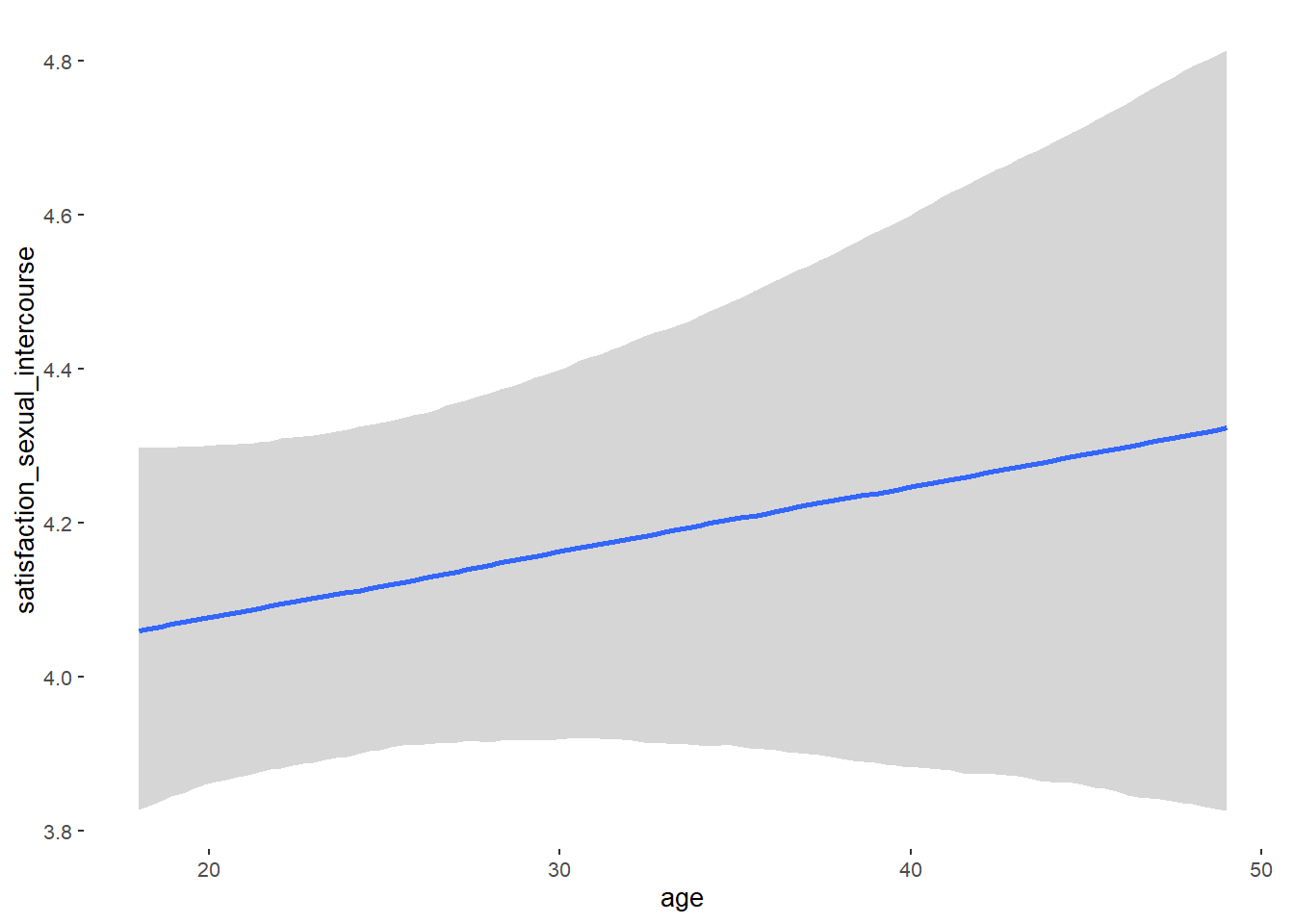
## age 0.01 0.01 -0.01 0.02 1.00 3400
## net_incomeeuro_500_1000 0.03 0.10 -0.14 0.19 1.00 2752
## net_incomeeuro_1000_2000 -0.08 0.13 -0.29 0.13 1.00 2490
## net_incomeeuro_2000_3000 -0.08 0.17 -0.37 0.20 1.00 2779
## net_incomeeuro_gt_3000 -0.26 0.29 -0.73 0.22 1.00 4165
## net_incomedont_tell -0.04 0.25 -0.46 0.36 1.00 3354
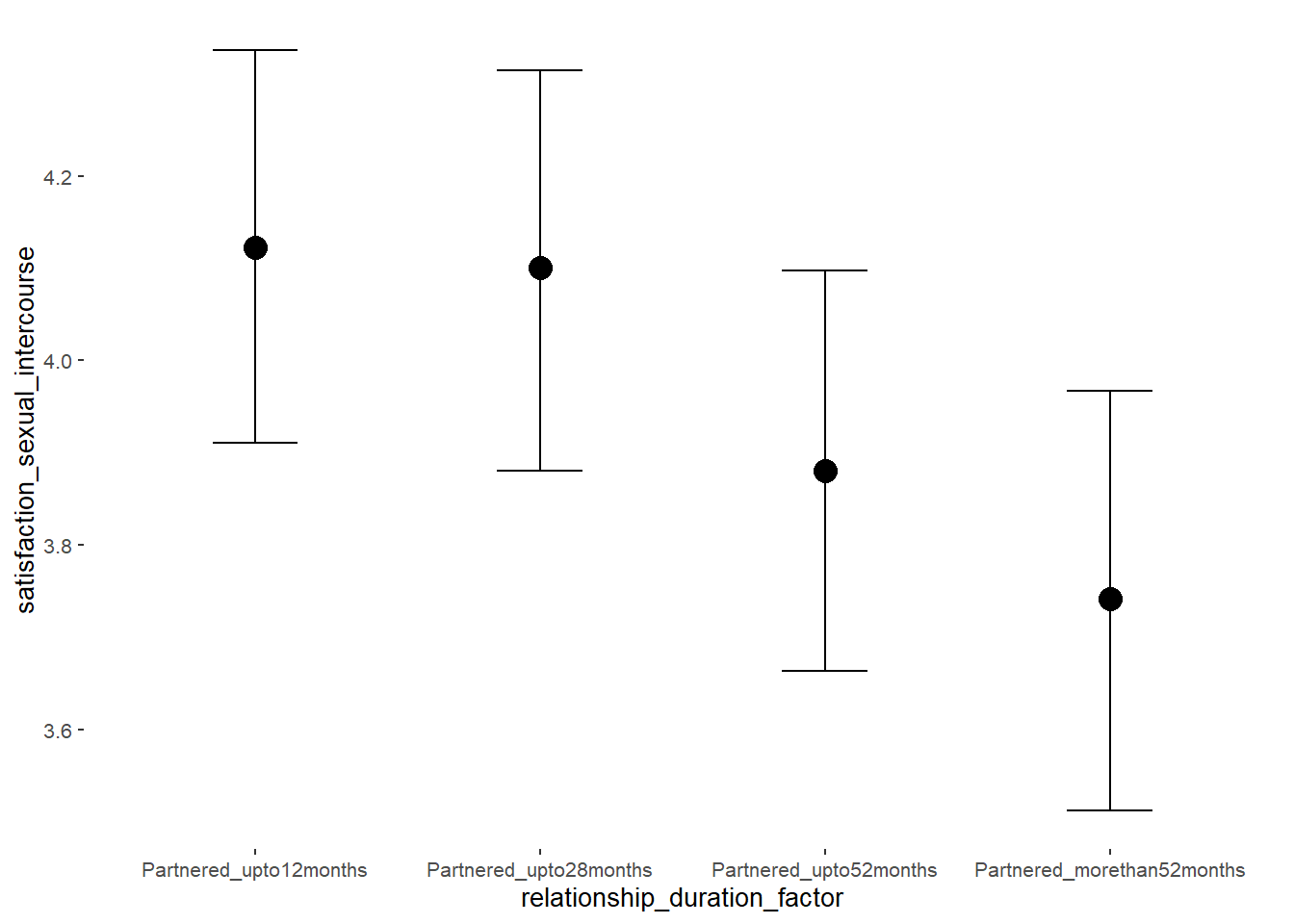
## relationship_duration_factorPartnered_upto28months -0.02 0.10 -0.19 0.15 1.00 3083
## relationship_duration_factorPartnered_upto52months -0.24 0.11 -0.42 -0.06 1.00 2824
## relationship_duration_factorPartnered_morethan52months -0.38 0.11 -0.57 -0.20 1.00 2557
## education_years -0.00 0.01 -0.02 0.01 1.00 5674
## bfi_extra 0.11 0.05 0.02 0.19 1.00 4252
## bfi_neuro -0.07 0.06 -0.16 0.03 1.00 4347
## bfi_agree 0.13 0.07 0.02 0.24 1.00 4543
## bfi_consc 0.14 0.06 0.04 0.23 1.00 4518
## bfi_open -0.10 0.06 -0.20 0.00 1.00 4654
## religiosity -0.01 0.03 -0.05 0.04 1.00 5221
## Tail_ESS
## Intercept 3486
## contraception_hormonalyes 3157
## age 3264
## net_incomeeuro_500_1000 2829
## net_incomeeuro_1000_2000 2906
## net_incomeeuro_2000_3000 3014
## net_incomeeuro_gt_3000 3219
## net_incomedont_tell 3052
## relationship_duration_factorPartnered_upto28months 3211
## relationship_duration_factorPartnered_upto52months 3394
## relationship_duration_factorPartnered_morethan52months 2923
## education_years 2946
## bfi_extra 3350
## bfi_neuro 3131
## bfi_agree 3177
## bfi_consc 3165
## bfi_open 3259
## religiosity 3199
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 1.03 0.03 0.99 1.07 1.00 5435 3222
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_hc_sexsat_controlled, range = c(-0.11, 0.11), ci = 0.90,
parameters = "contraception"))## Picking joint bandwidth of 0.0111## Warning: Removed 399 rows containing non-finite values (stat_density_ridges).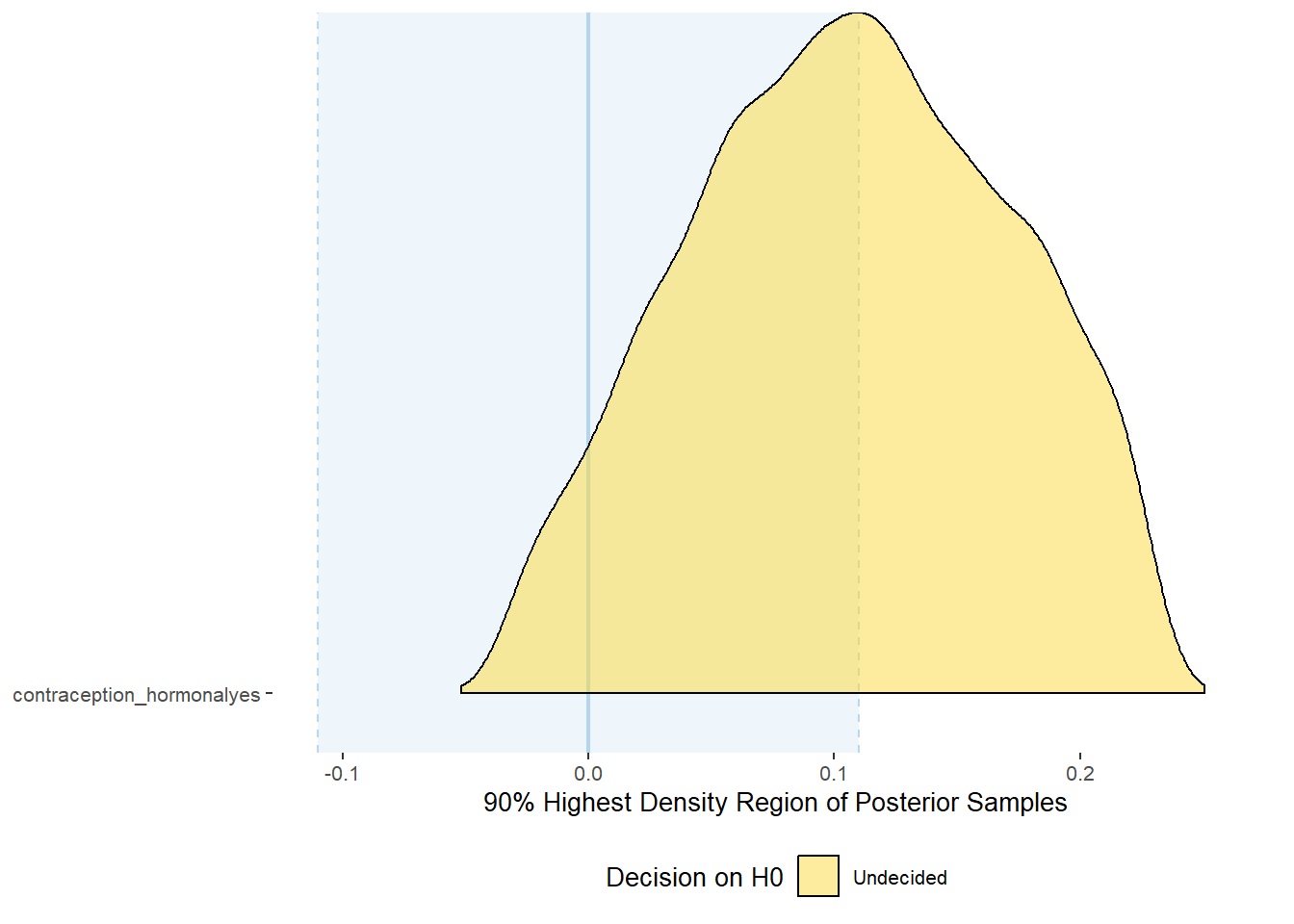
equivalence_test(m_hc_sexsat_controlled, range = c(-0.11, 0.11), ci = 0.90,
parameters = "contraception")## # A tibble: 1 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.11 0.11 0.518 Undecided -0.0319 0.230 fixed conditio~Forest Plot for Effect Sizes
m_hc_sexsat_controlled %>%
spread_draws(b_contraception_hormonalyes,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity) %>%
pivot_longer(cols = c(b_contraception_hormonalyes,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_relationship_duration_factor",
"Relationship Duration",
ifelse(condition %contains% "b_net_income",
"Income",
NA)),
group = ifelse(condition %contains% "b_contraception_hormonalyes",
"Contraception", group),
condition = ifelse(condition %contains% "b_contraception_hormonalyes",
"Hormonal Contraception", condition),
condition = ifelse(condition == "b_age", "Age",
ifelse(condition == "b_net_incomeeuro_500_1000", "500-1000 Euro",
ifelse(condition == "b_net_incomeeuro_1000_2000", "1000-2000 Euro",
ifelse(condition == "b_net_incomeeuro_2000_3000", "2000-3000 Euro",
ifelse(condition == "b_net_incomeeuro_gt_3000", ">3000 Euro",
ifelse(condition == "b_net_incomedont_tell", "do not tell",
ifelse(condition == "b_relationship_duration_factorPartnered_upto28months",
"13-28 months",
ifelse(condition == "b_relationship_duration_factorPartnered_upto52months",
"29-52 months",
ifelse(condition == "b_relationship_duration_factorPartnered_morethan52months",
">52 months",
ifelse(condition == "b_education_years", "Years of Education",
ifelse(condition == "b_bfi_extra", "Extraversion",
ifelse(condition == "b_bfi_neuro", "Neuroticism",
ifelse(condition == "b_bfi_agree", "Agreeableness",
ifelse(condition == "b_bfi_consc", "Conscientiousness",
ifelse(condition == "b_bfi_open", "Openness",
ifelse(condition == "b_religiosity", "Religiosity",
condition)))))))))))))))),
group = ifelse(is.na(group), condition, group),
condition = factor(condition, levels = rev(c("Hormonal Contraception", "Age",
"500-1000 Euro", "1000-2000 Euro",
"2000-3000 Euro", ">3000 Euro", "do not tell",
"13-28 months", "29-52 months",
">52 months",
"Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))),
group = factor(group, levels = c("Contraception", "Age", "Income",
"Relationship Duration","Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.11))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.11, 0.11), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")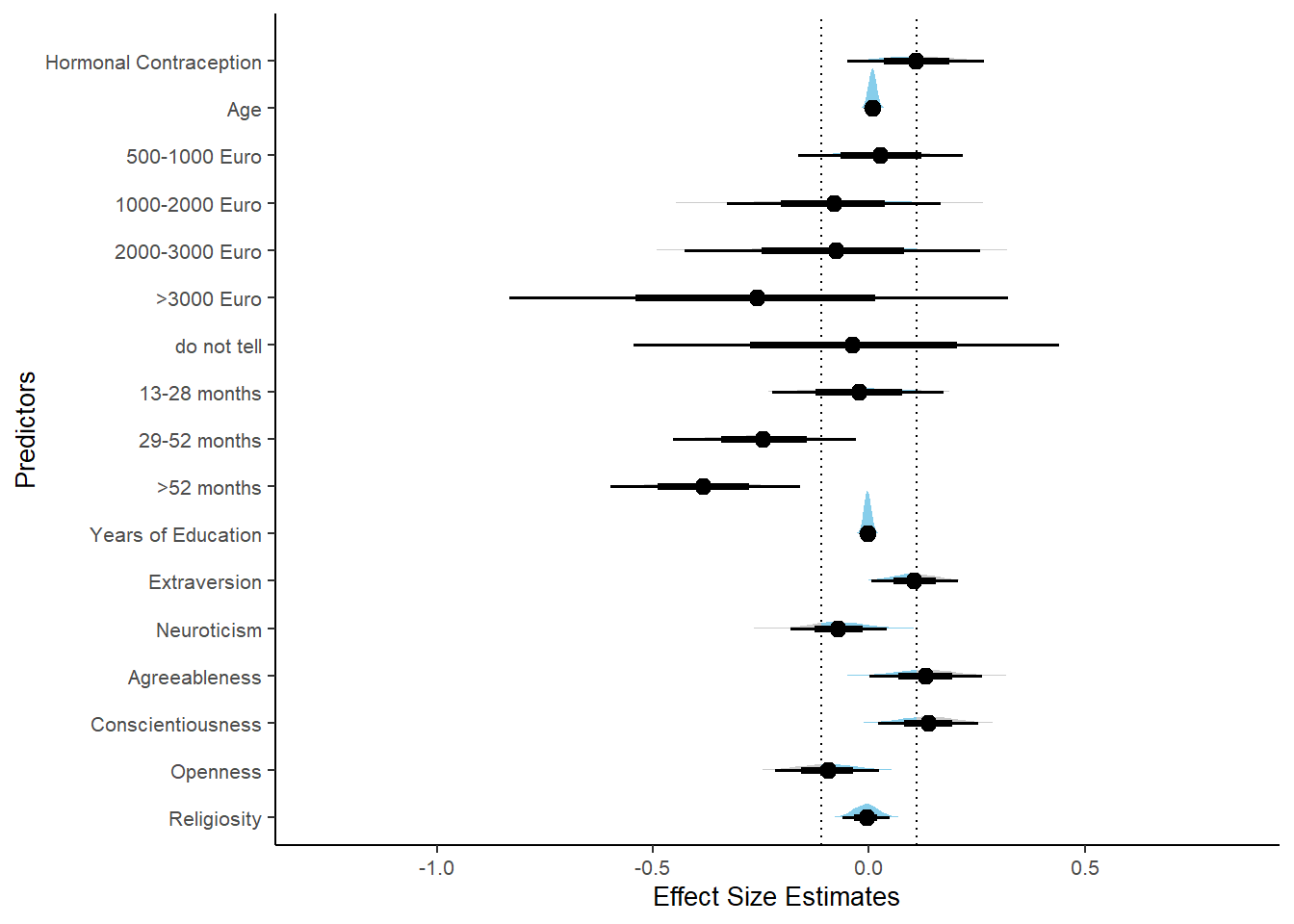
Libido
Model
Summary
## Family: gaussian
## Links: mu = identity; sigma = identity
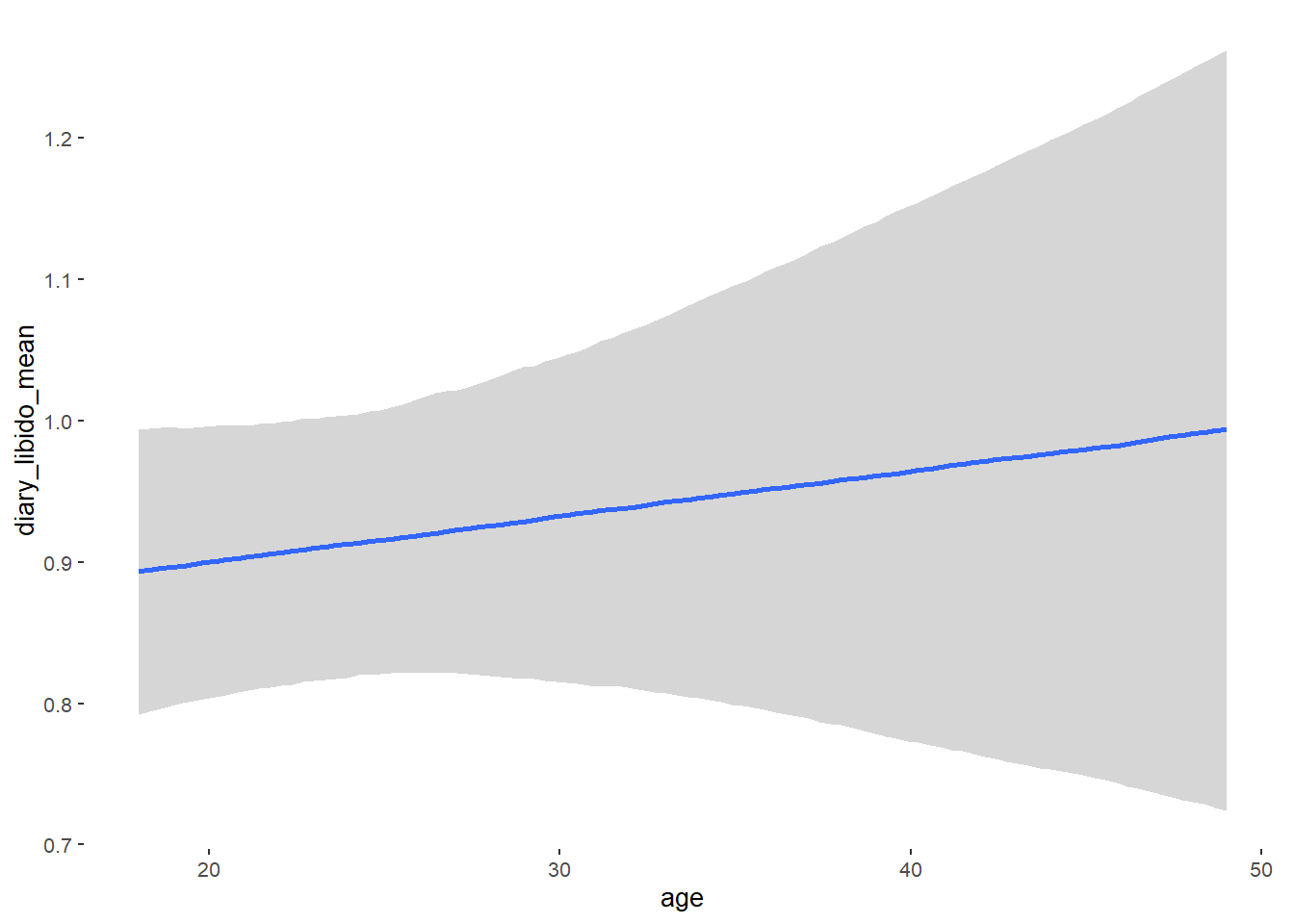
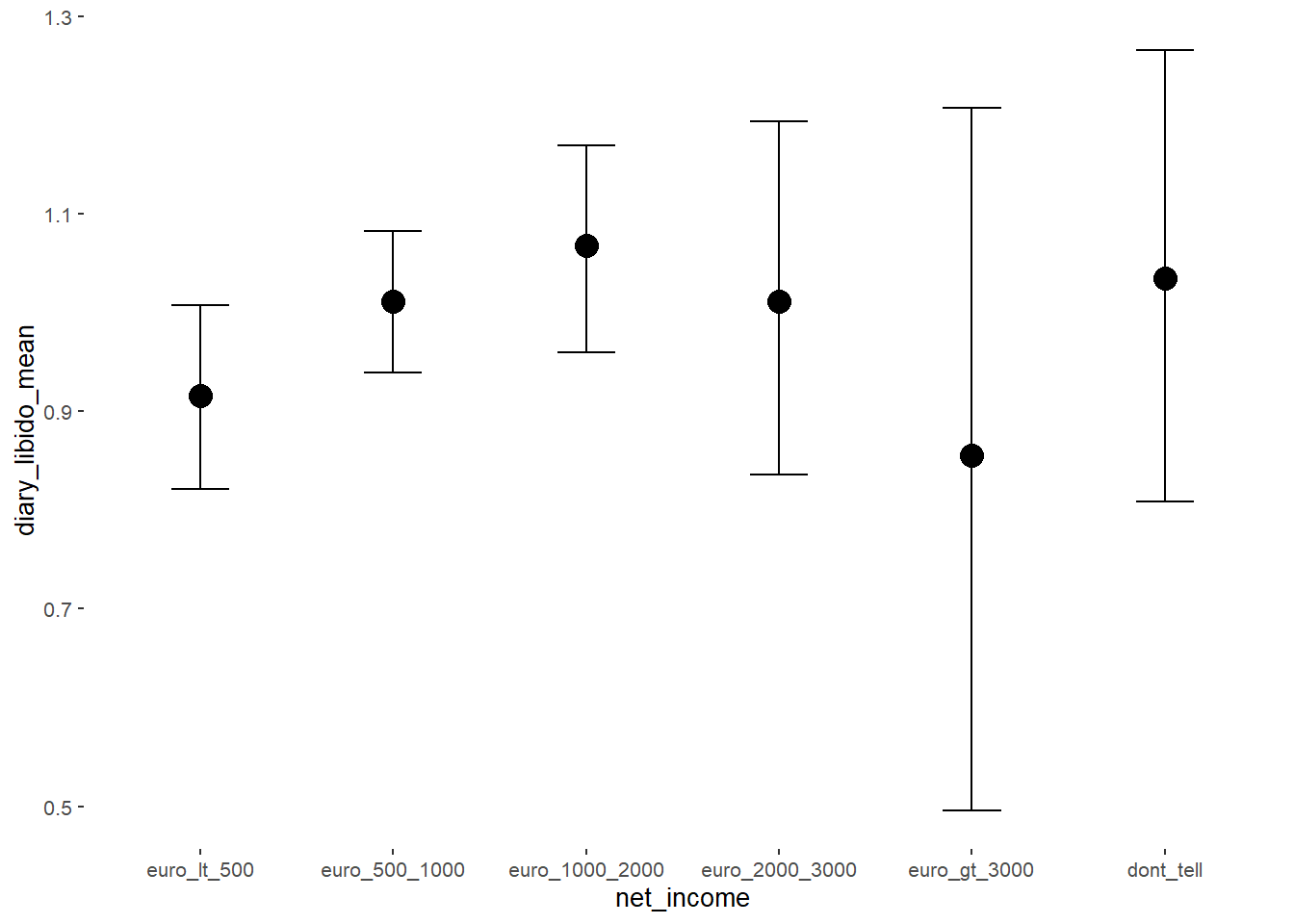
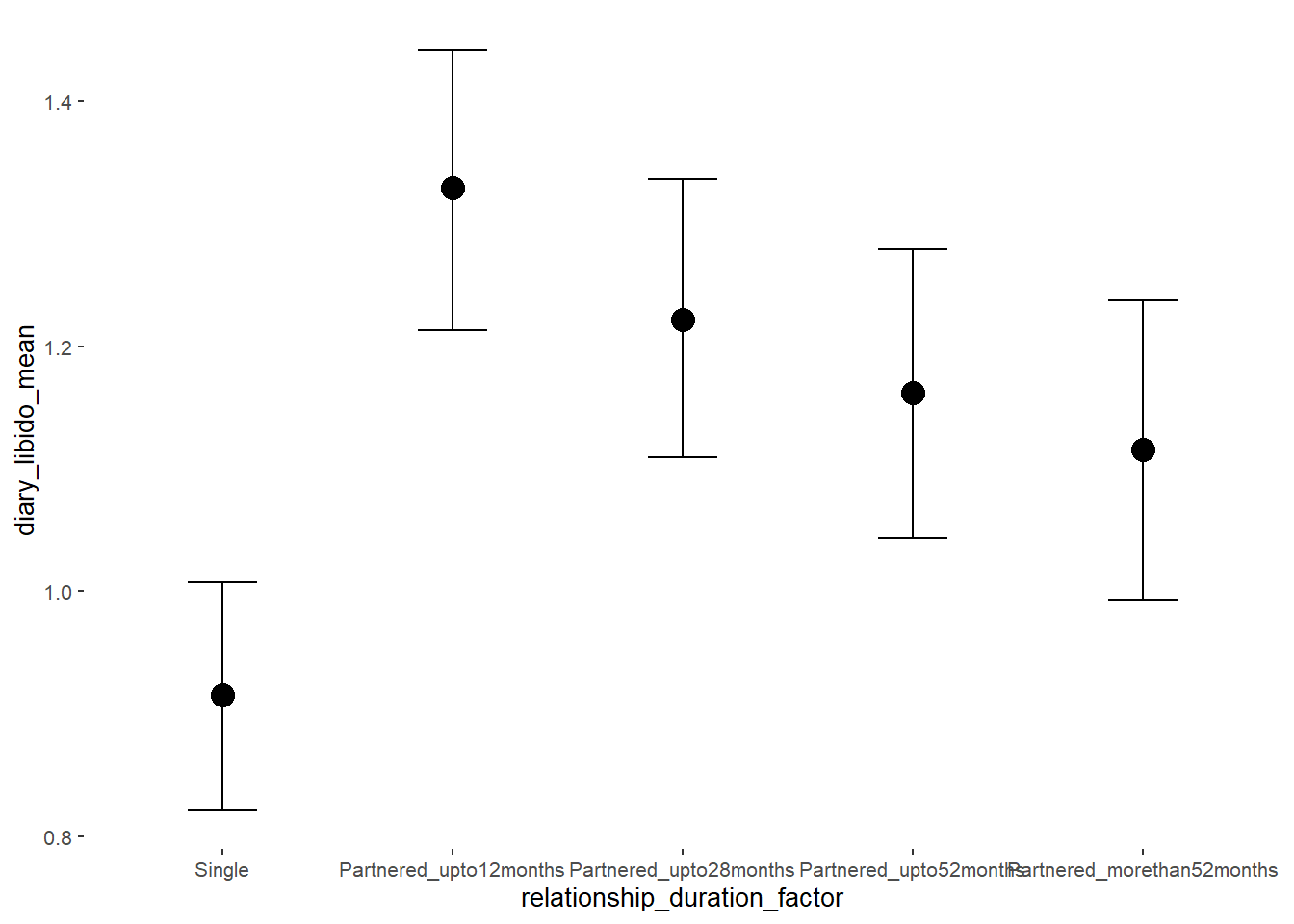
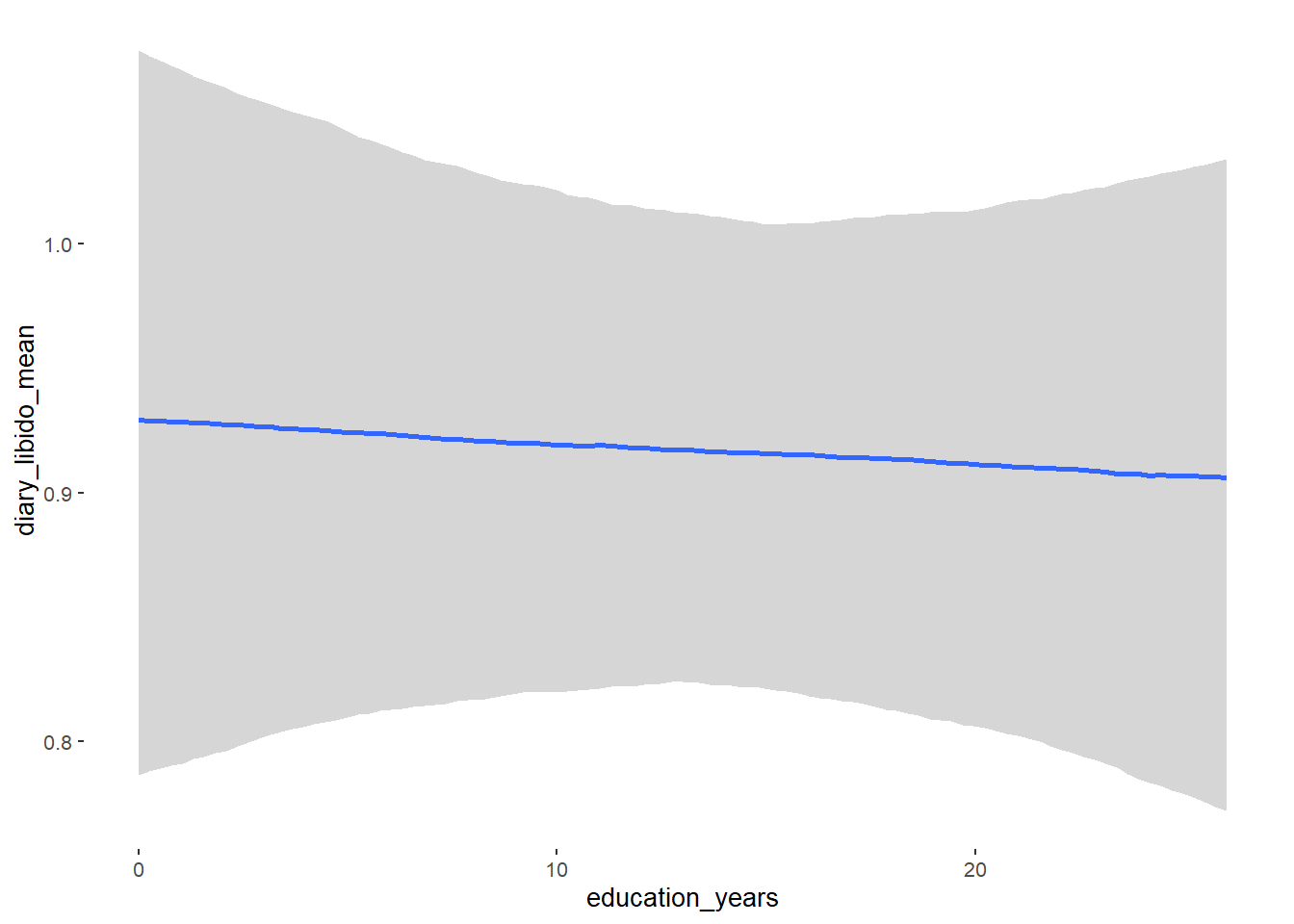
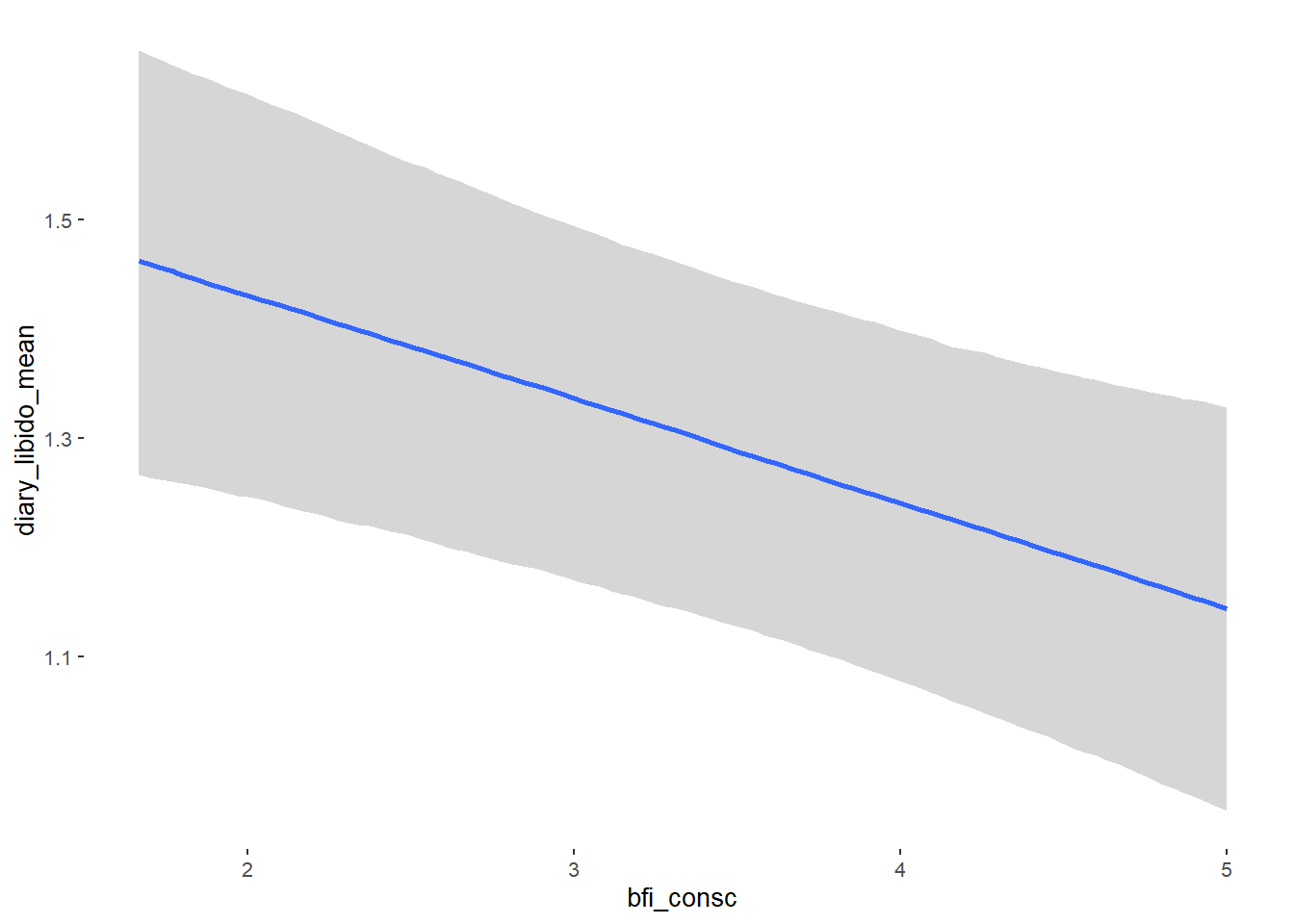
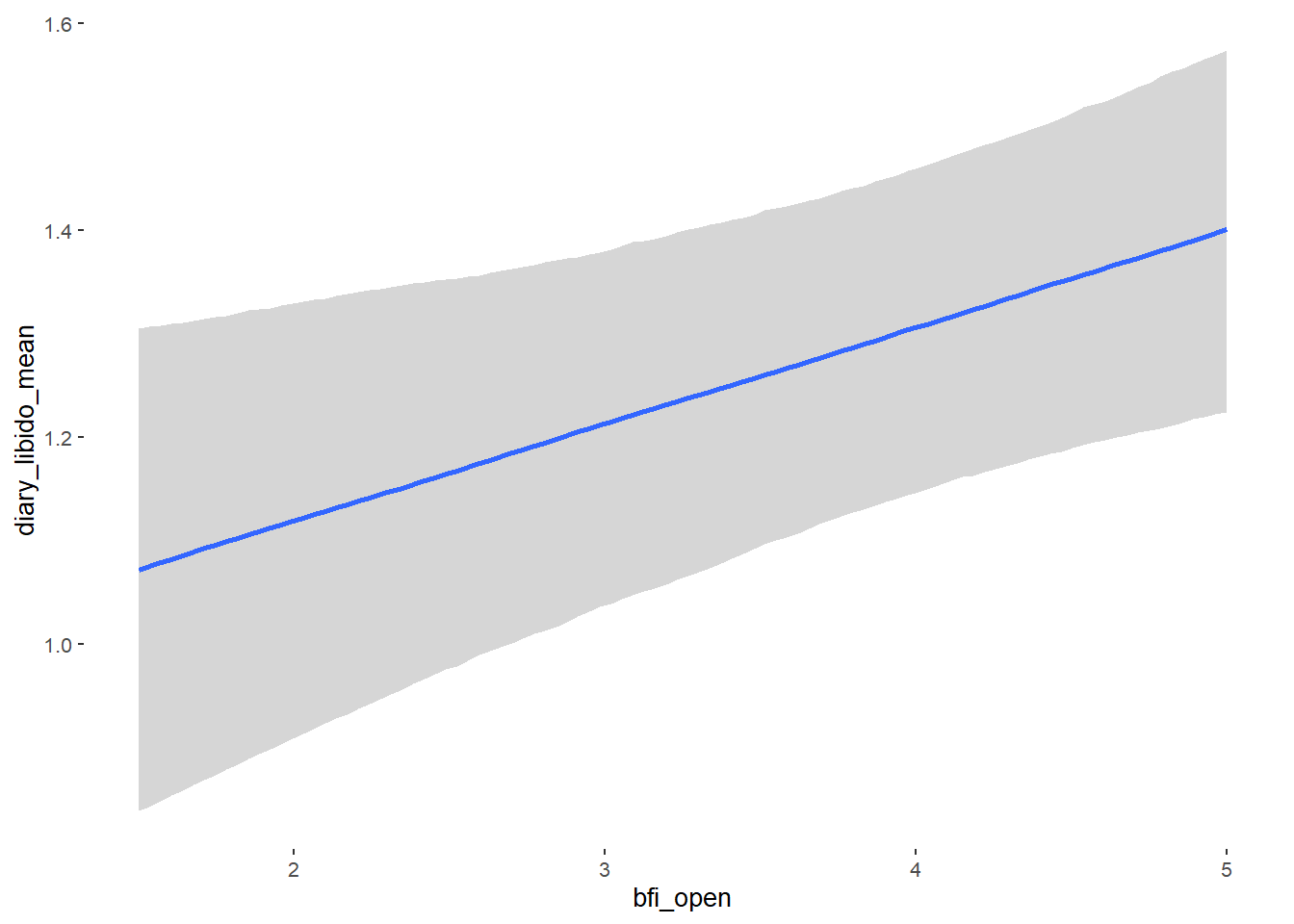
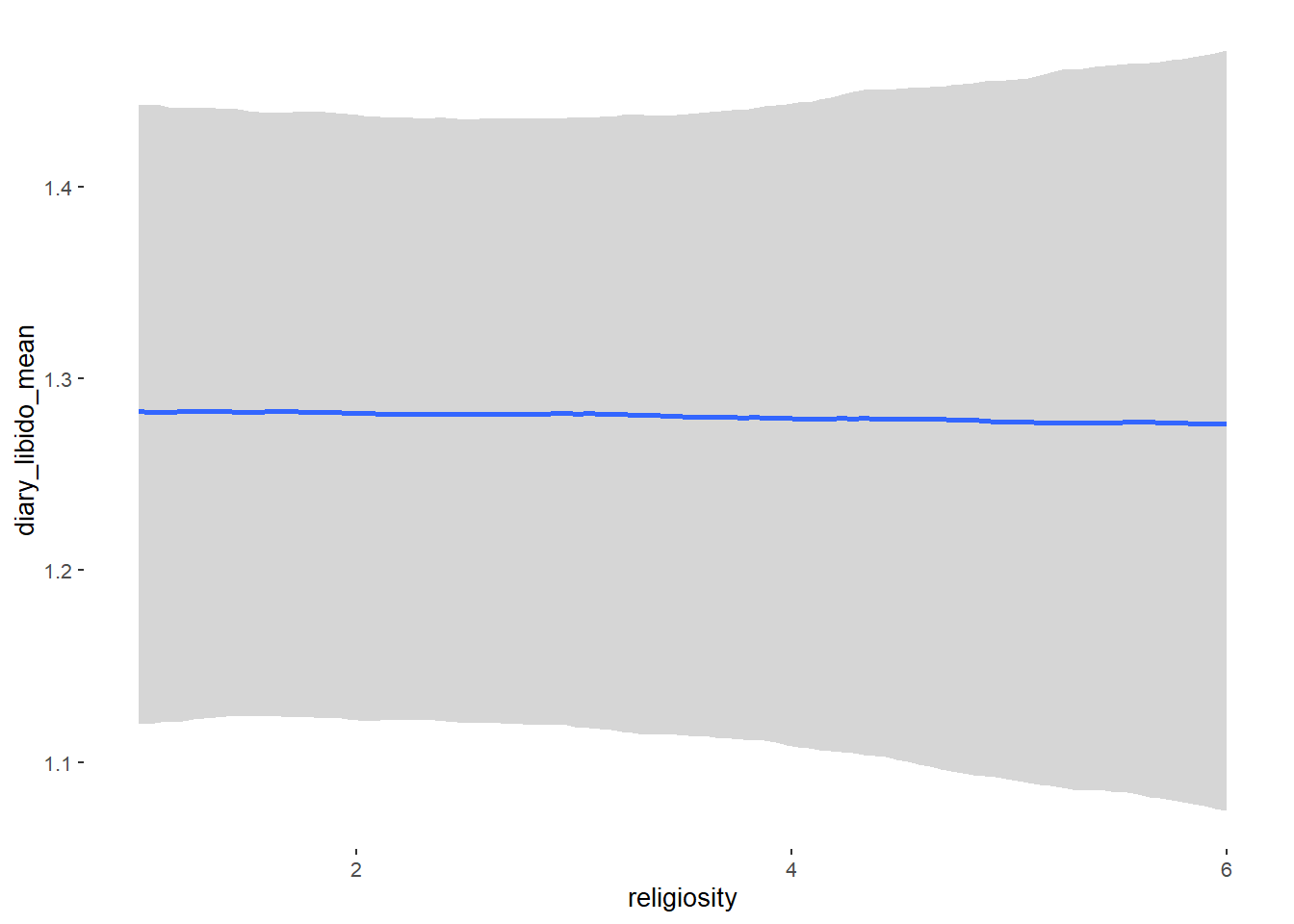
## Formula: diary_libido_mean ~ contraception_hormonal + age + net_income + relationship_duration_factor + education_years + bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open + religiosity
## Data: data (Number of observations: 968)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept 0.27 0.26 -0.15 0.70 1.00 4585
## contraception_hormonalyes 0.00 0.04 -0.06 0.07 1.00 4109
## age 0.00 0.00 -0.00 0.01 1.00 3543
## net_incomeeuro_500_1000 0.10 0.05 0.02 0.17 1.00 2903
## net_incomeeuro_1000_2000 0.15 0.06 0.04 0.26 1.00 2443
## net_incomeeuro_2000_3000 0.10 0.10 -0.06 0.26 1.00 3429
## net_incomeeuro_gt_3000 -0.06 0.18 -0.35 0.24 1.00 4216
## net_incomedont_tell 0.12 0.12 -0.08 0.33 1.00 3409
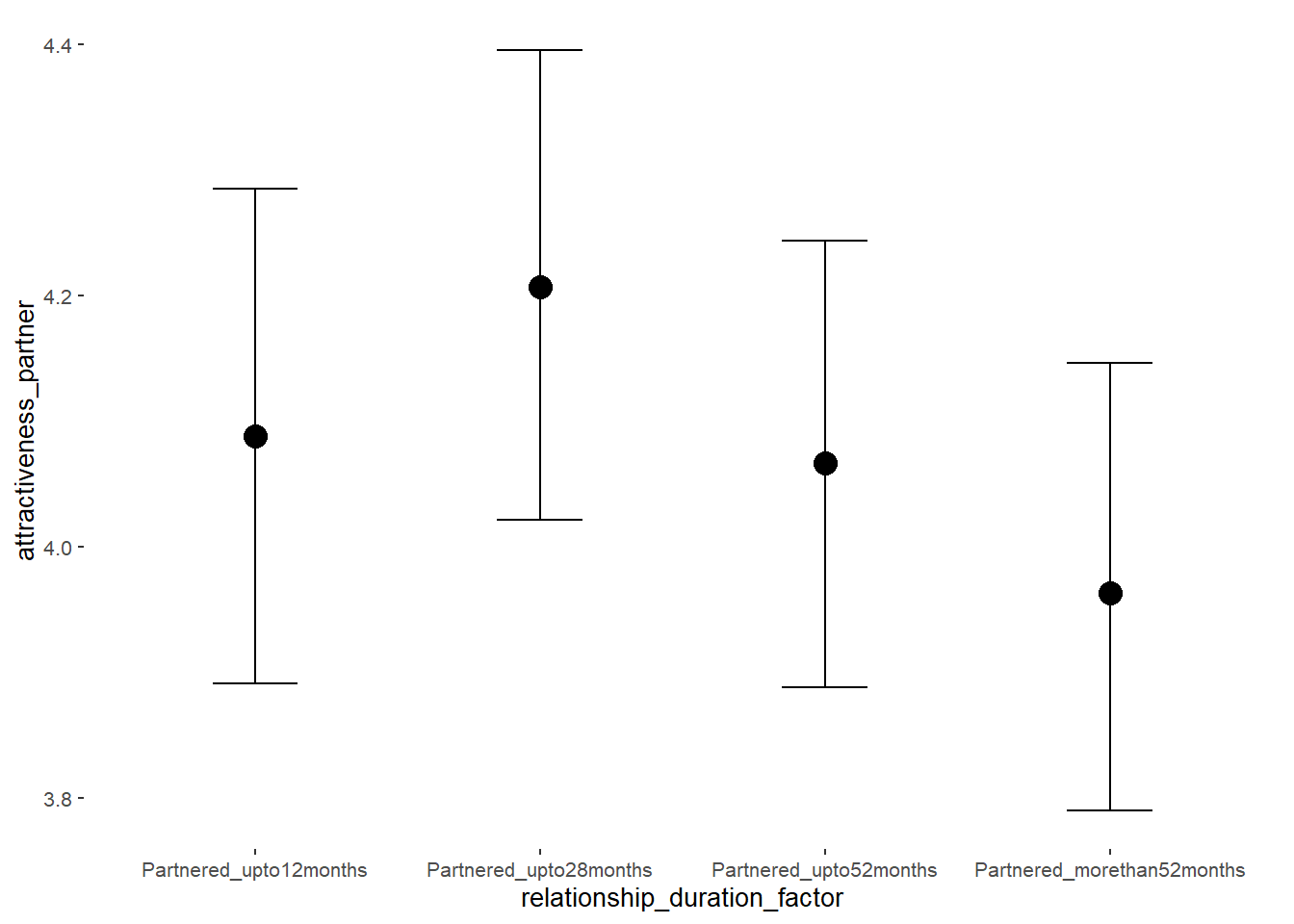
## relationship_duration_factorPartnered_upto12months 0.41 0.05 0.32 0.50 1.00 3379
## relationship_duration_factorPartnered_upto28months 0.31 0.05 0.22 0.39 1.00 3316
## relationship_duration_factorPartnered_upto52months 0.25 0.06 0.15 0.34 1.00 3128
## relationship_duration_factorPartnered_morethan52months 0.20 0.06 0.10 0.30 1.00 3302
## education_years -0.00 0.00 -0.01 0.01 1.00 6409
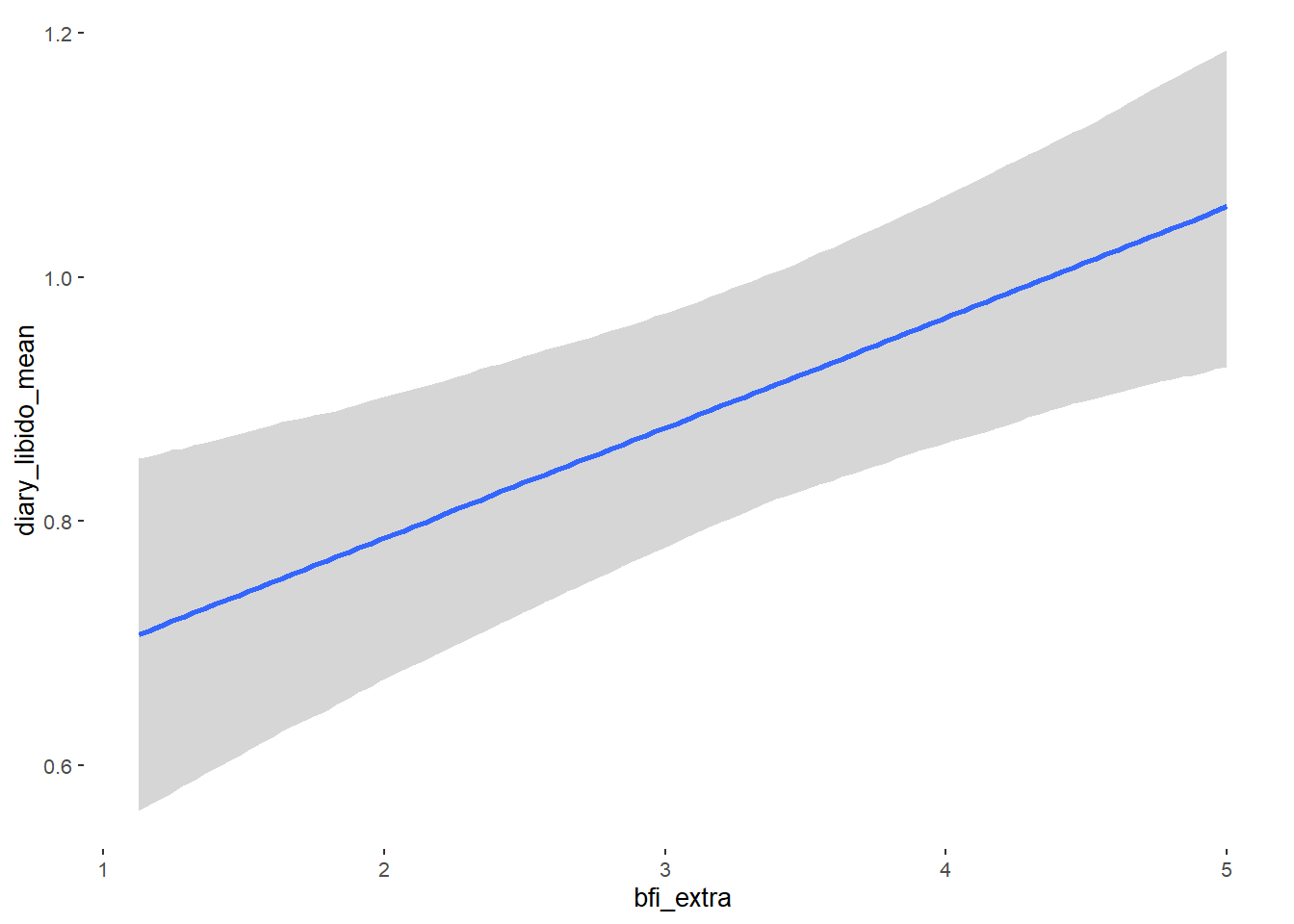
## bfi_extra 0.09 0.03 0.05 0.13 1.00 4187
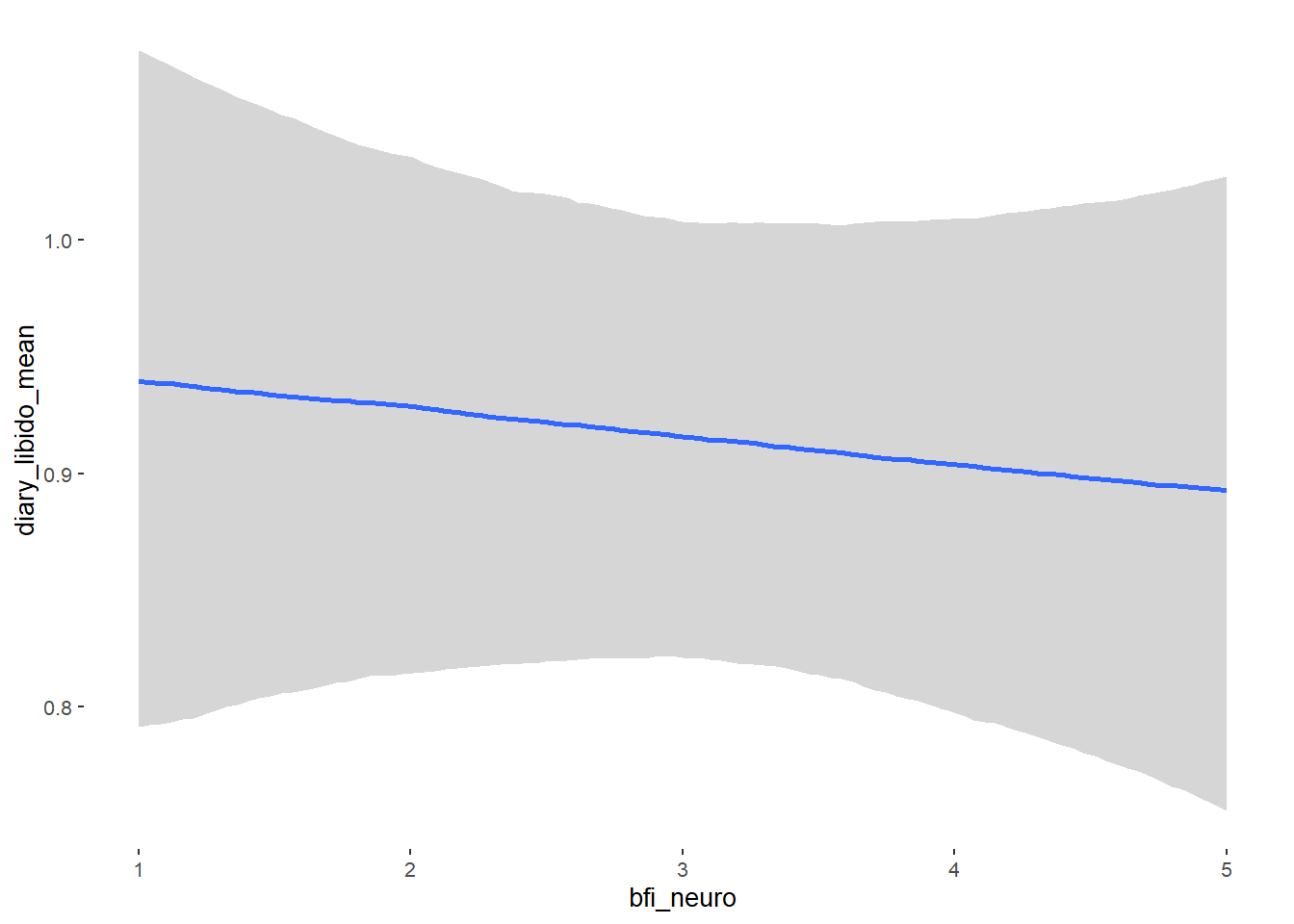
## bfi_neuro -0.01 0.03 -0.06 0.03 1.00 4280
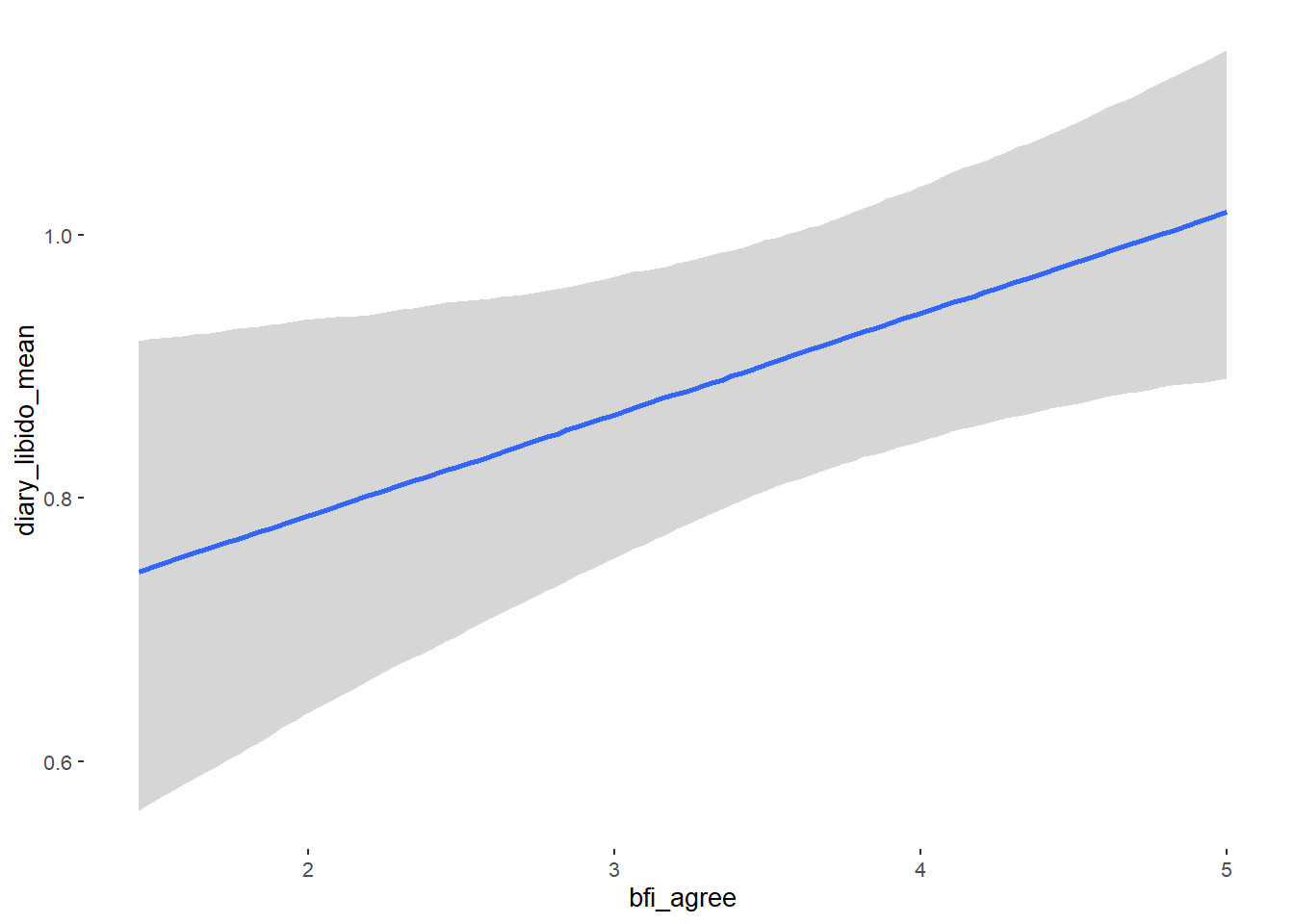
## bfi_agree 0.08 0.03 0.02 0.13 1.00 4350
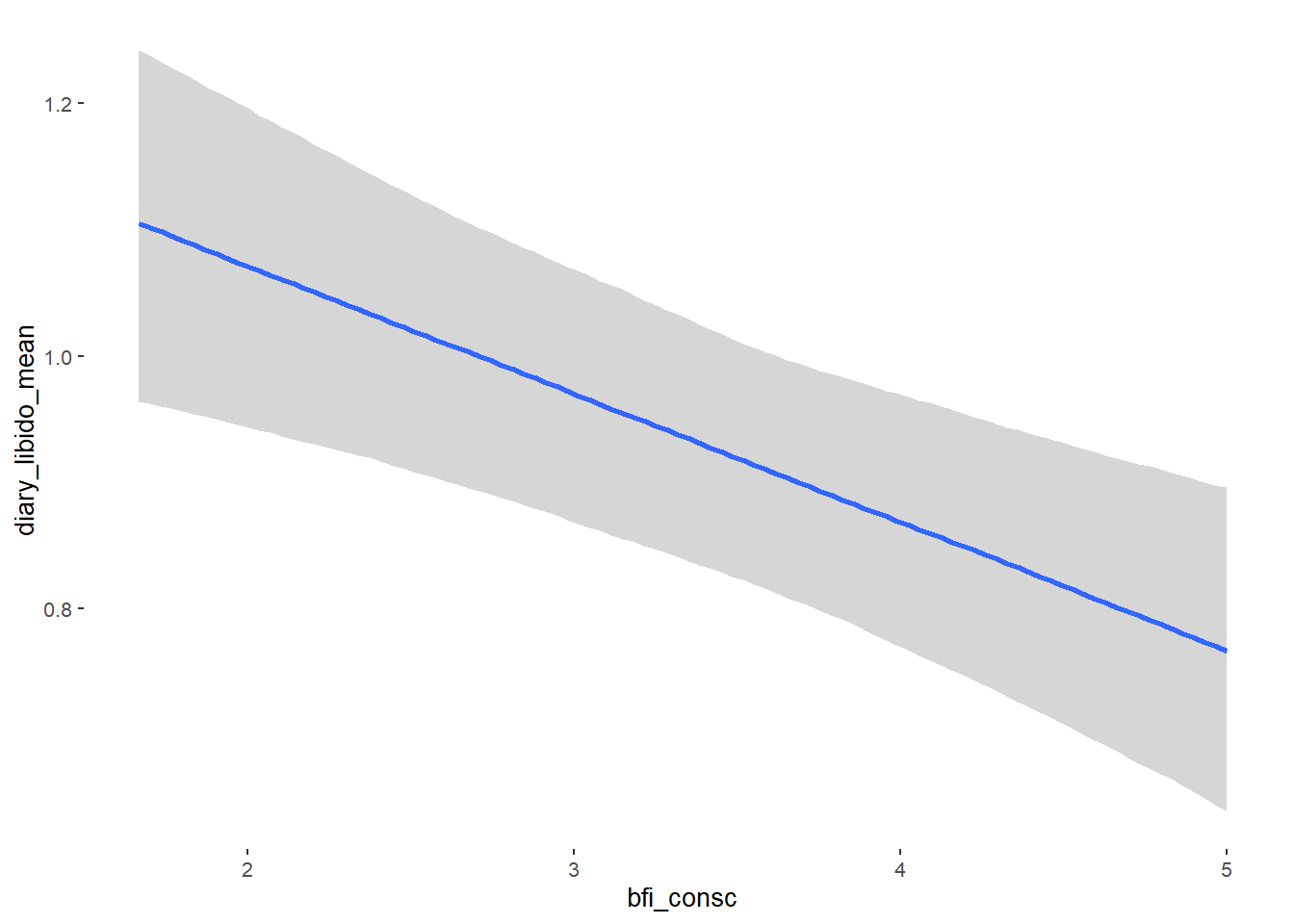
## bfi_consc -0.10 0.03 -0.15 -0.05 1.00 4833
## bfi_open 0.10 0.03 0.05 0.15 1.00 3759
## religiosity -0.01 0.01 -0.03 0.02 1.00 5616
## Tail_ESS
## Intercept 3321
## contraception_hormonalyes 3244
## age 3209
## net_incomeeuro_500_1000 3187
## net_incomeeuro_1000_2000 3087
## net_incomeeuro_2000_3000 2672
## net_incomeeuro_gt_3000 2636
## net_incomedont_tell 3193
## relationship_duration_factorPartnered_upto12months 3115
## relationship_duration_factorPartnered_upto28months 3018
## relationship_duration_factorPartnered_upto52months 2549
## relationship_duration_factorPartnered_morethan52months 2786
## education_years 3125
## bfi_extra 3272
## bfi_neuro 3392
## bfi_agree 3264
## bfi_consc 3278
## bfi_open 2798
## religiosity 2937
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.56 0.01 0.54 0.58 1.00 5416 3078
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_hc_libido_controlled, range = c(-0.06, 0.06), ci = 0.90,
parameters = "contraception"))## Picking joint bandwidth of 0.00559## Warning: Removed 399 rows containing non-finite values (stat_density_ridges).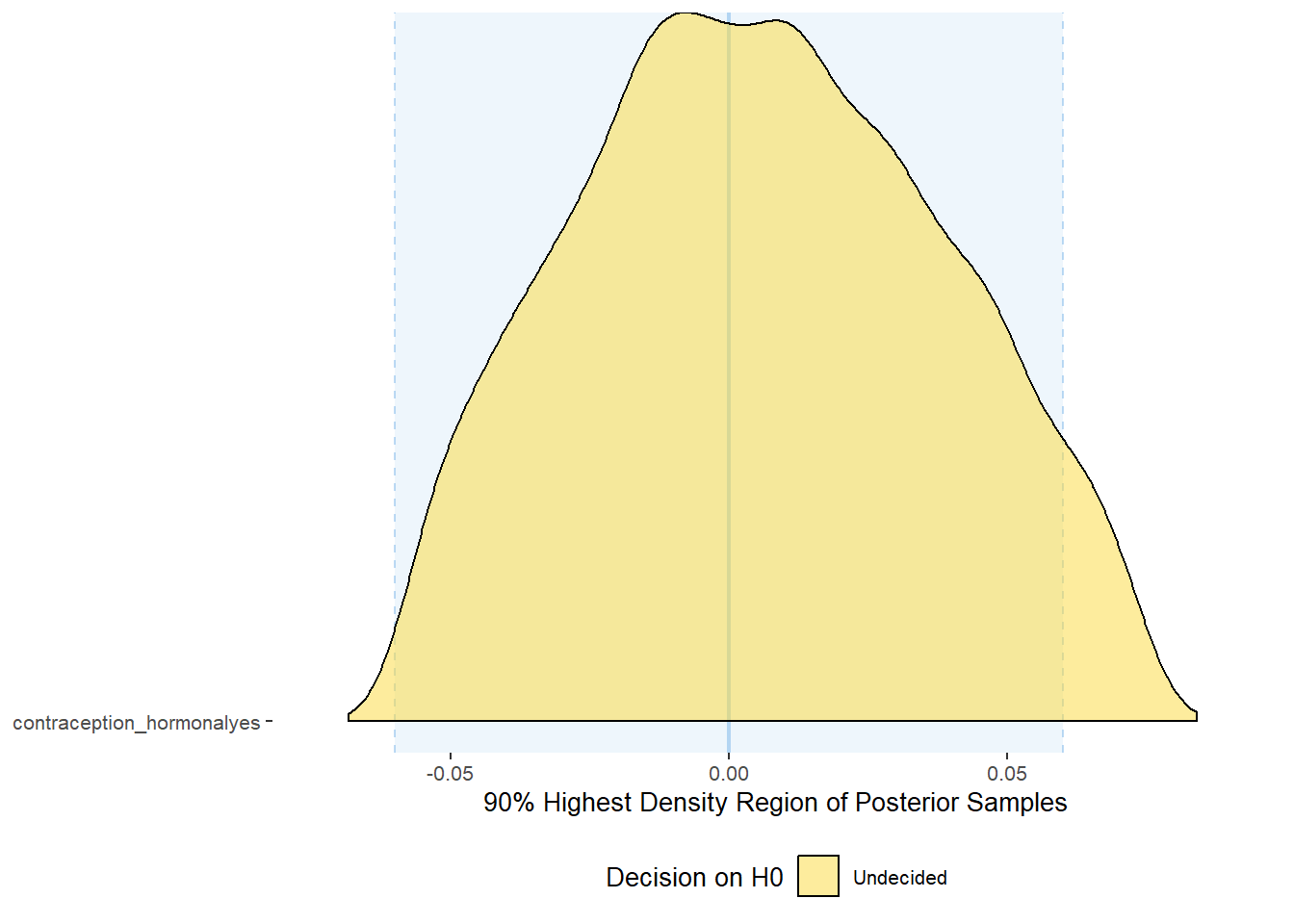
equivalence_test(m_hc_libido_controlled, range = c(-0.06, 0.06), ci = 0.90,
parameters = "contraception")## # A tibble: 1 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.06 0.06 0.949 Undecided -0.0570 0.0741 fixed conditio~Forest Plot for Effect Sizes
m_hc_libido_controlled %>%
spread_draws(b_contraception_hormonalyes,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity) %>%
pivot_longer(cols = c(b_contraception_hormonalyes,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_relationship_duration_factor",
"Relationship Duration",
ifelse(condition %contains% "b_net_income",
"Income",
NA)),
group = ifelse(condition %contains% "b_contraception_hormonalyes",
"Contraception", group),
condition = ifelse(condition %contains% "b_contraception_hormonalyes",
"Hormonal Contraception", condition),
condition = ifelse(condition == "b_age", "Age",
ifelse(condition == "b_net_incomeeuro_500_1000", "500-1000 Euro",
ifelse(condition == "b_net_incomeeuro_1000_2000", "1000-2000 Euro",
ifelse(condition == "b_net_incomeeuro_2000_3000", "2000-3000 Euro",
ifelse(condition == "b_net_incomeeuro_gt_3000", ">3000 Euro",
ifelse(condition == "b_net_incomedont_tell", "do not tell",
ifelse(condition == "b_relationship_duration_factorPartnered_upto28months",
"13-28 months",
ifelse(condition == "b_relationship_duration_factorPartnered_upto52months",
"29-52 months",
ifelse(condition == "b_relationship_duration_factorPartnered_morethan52months",
">52 months",
ifelse(condition == "b_education_years", "Years of Education",
ifelse(condition == "b_bfi_extra", "Extraversion",
ifelse(condition == "b_bfi_neuro", "Neuroticism",
ifelse(condition == "b_bfi_agree", "Agreeableness",
ifelse(condition == "b_bfi_consc", "Conscientiousness",
ifelse(condition == "b_bfi_open", "Openness",
ifelse(condition == "b_religiosity", "Religiosity",
condition)))))))))))))))),
group = ifelse(is.na(group), condition, group),
condition = factor(condition, levels = rev(c("Hormonal Contraception", "Age",
"500-1000 Euro", "1000-2000 Euro",
"2000-3000 Euro", ">3000 Euro", "do not tell",
"13-28 months", "29-52 months",
">52 months",
"Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))),
group = factor(group, levels = c("Contraception", "Age", "Income",
"Relationship Duration","Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.06))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.06, 0.06), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")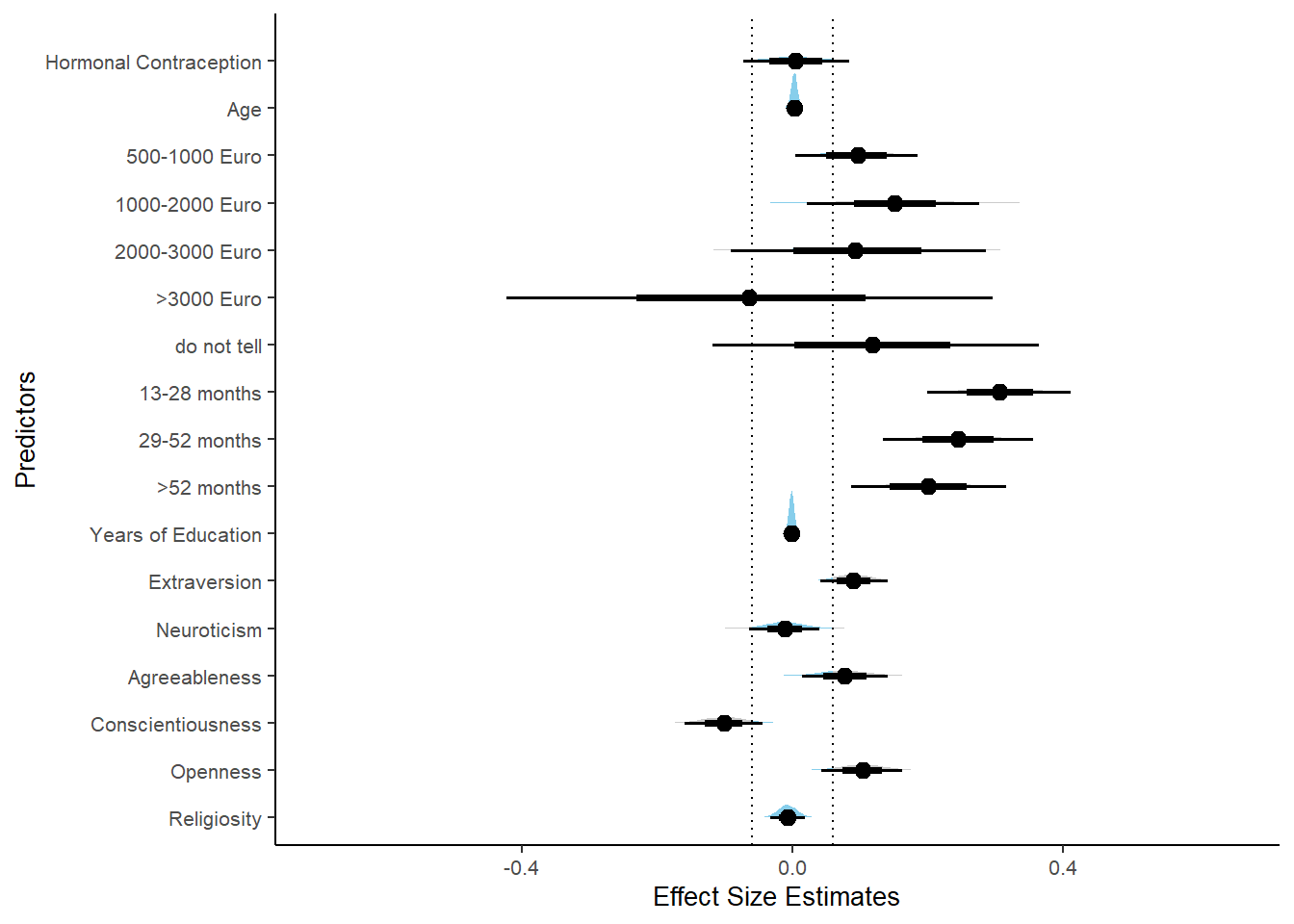
Sexual Frequency (Penetrative Intercourse)
Model
m_hc_sexfreqpen_controlled = brm(diary_sex_active_sex_sum ~
offset(log(number_of_days)) +
contraception_hormonal +
age + net_income + relationship_duration_factor +
education_years +
bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open +
religiosity,
data = data, family = poisson(),
file = "m_hc_sexfreqpen_controlled")Summary
## Family: poisson
## Links: mu = log
## Formula: diary_sex_active_sex_sum ~ offset(log(number_of_days)) + contraception_hormonal + age + net_income + relationship_duration_factor + education_years + bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open + religiosity
## Data: data (Number of observations: 897)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept -3.35 0.18 -3.64 -3.06 1.00 4112
## contraception_hormonalyes 0.16 0.03 0.12 0.21 1.00 4523
## age -0.00 0.00 -0.01 0.01 1.00 3788
## net_incomeeuro_500_1000 0.16 0.03 0.11 0.21 1.00 2897
## net_incomeeuro_1000_2000 0.20 0.04 0.12 0.27 1.00 2694
## net_incomeeuro_2000_3000 0.38 0.06 0.28 0.47 1.00 2844
## net_incomeeuro_gt_3000 0.05 0.12 -0.14 0.24 1.00 3458
## net_incomedont_tell 0.45 0.07 0.32 0.56 1.00 3002
## relationship_duration_factorPartnered_upto12months 1.34 0.04 1.27 1.41 1.00 2312
## relationship_duration_factorPartnered_upto28months 1.22 0.04 1.15 1.29 1.00 2162
## relationship_duration_factorPartnered_upto52months 0.96 0.05 0.88 1.04 1.00 2430
## relationship_duration_factorPartnered_morethan52months 0.92 0.05 0.84 1.00 1.00 2440
## education_years -0.01 0.00 -0.01 -0.01 1.00 6844
## bfi_extra 0.01 0.02 -0.02 0.04 1.00 4316
## bfi_neuro -0.00 0.02 -0.03 0.03 1.00 3095
## bfi_agree 0.09 0.02 0.06 0.13 1.00 4297
## bfi_consc -0.02 0.02 -0.05 0.02 1.00 3935
## bfi_open 0.03 0.02 -0.00 0.06 1.00 3992
## religiosity -0.01 0.01 -0.03 0.00 1.00 4416
## Tail_ESS
## Intercept 3113
## contraception_hormonalyes 2707
## age 3007
## net_incomeeuro_500_1000 3055
## net_incomeeuro_1000_2000 2973
## net_incomeeuro_2000_3000 2933
## net_incomeeuro_gt_3000 3126
## net_incomedont_tell 3336
## relationship_duration_factorPartnered_upto12months 2836
## relationship_duration_factorPartnered_upto28months 2620
## relationship_duration_factorPartnered_upto52months 2665
## relationship_duration_factorPartnered_morethan52months 2728
## education_years 3286
## bfi_extra 2890
## bfi_neuro 3132
## bfi_agree 2658
## bfi_consc 2976
## bfi_open 3136
## religiosity 2969
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_hc_sexfreqpen_controlled, range = c(-0.05, 0.05), ci = 0.90,
parameters = "contraception"))## Picking joint bandwidth of 0.00371## Warning: Removed 399 rows containing non-finite values (stat_density_ridges).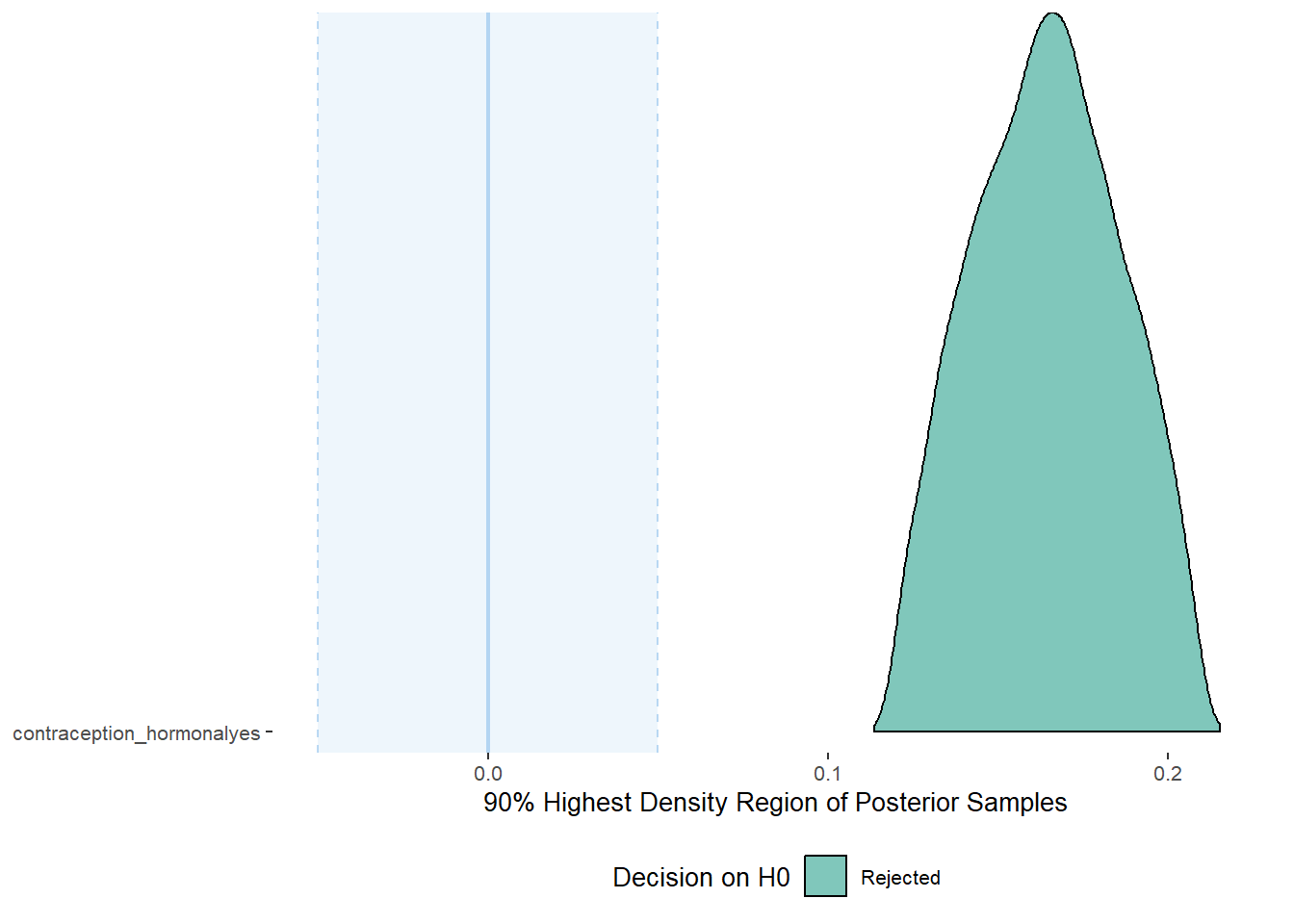
equivalence_test(m_hc_sexfreqpen_controlled, range = c(-0.05, 0.05), ci = 0.90,
parameters = "contraception")## # A tibble: 1 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.05 0.05 0 Rejected 0.120 0.209 fixed conditio~Plots
conditional_effects(m_hc_sexfreqpen_controlled,
effects = "contraception_hormonal",
conditions = data.frame(number_of_days = 1))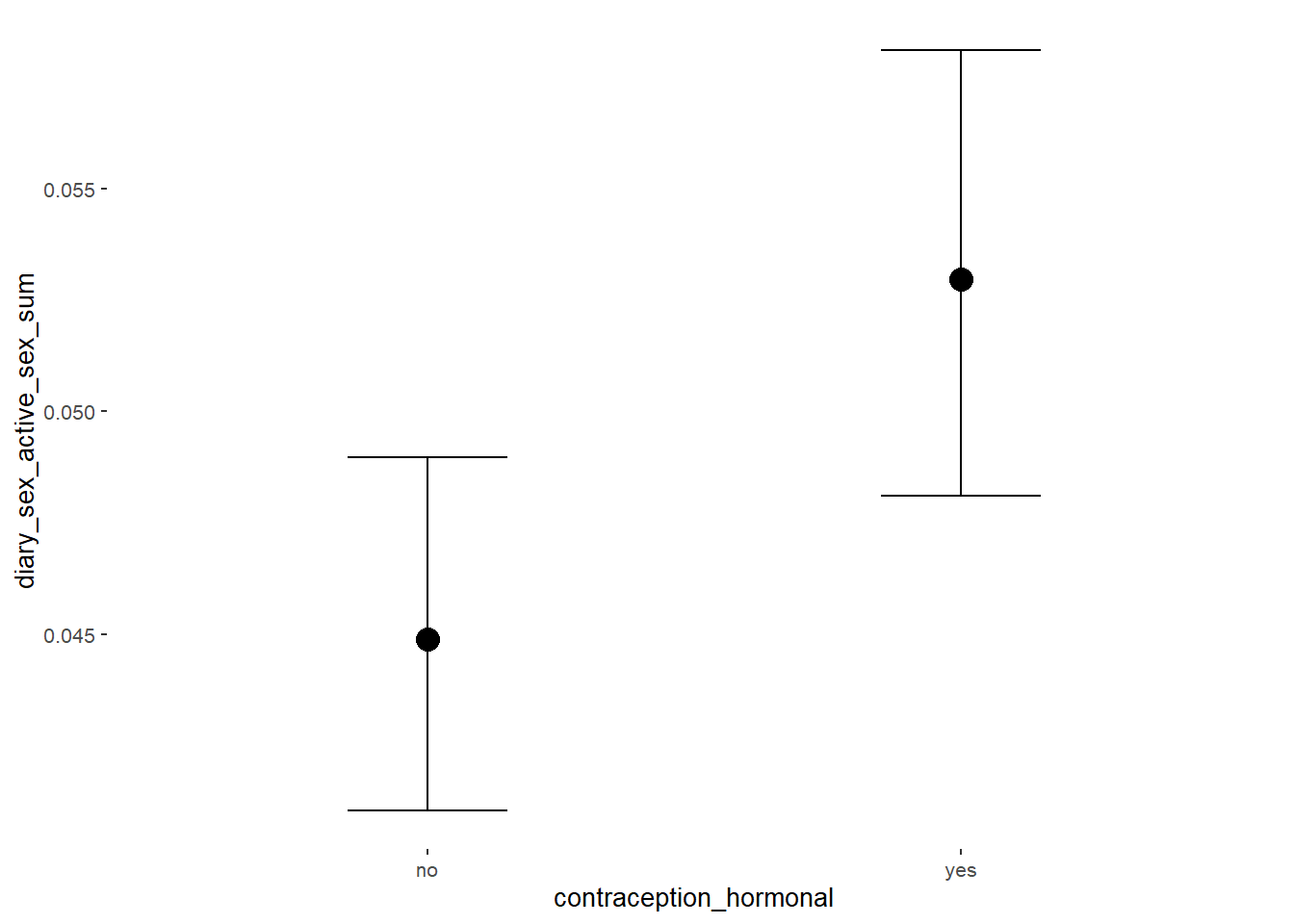
Forest Plot for Effect Sizes
m_hc_sexfreqpen_controlled %>%
spread_draws(b_contraception_hormonalyes,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity) %>%
pivot_longer(cols = c(b_contraception_hormonalyes,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_relationship_duration_factor",
"Relationship Duration",
ifelse(condition %contains% "b_net_income",
"Income",
NA)),
group = ifelse(condition %contains% "b_contraception_hormonalyes",
"Contraception", group),
condition = ifelse(condition %contains% "b_contraception_hormonalyes",
"Hormonal Contraception", condition),
condition = ifelse(condition == "b_age", "Age",
ifelse(condition == "b_net_incomeeuro_500_1000", "500-1000 Euro",
ifelse(condition == "b_net_incomeeuro_1000_2000", "1000-2000 Euro",
ifelse(condition == "b_net_incomeeuro_2000_3000", "2000-3000 Euro",
ifelse(condition == "b_net_incomeeuro_gt_3000", ">3000 Euro",
ifelse(condition == "b_net_incomedont_tell", "do not tell",
ifelse(condition == "b_relationship_duration_factorPartnered_upto28months",
"13-28 months",
ifelse(condition == "b_relationship_duration_factorPartnered_upto52months",
"29-52 months",
ifelse(condition == "b_relationship_duration_factorPartnered_morethan52months",
">52 months",
ifelse(condition == "b_education_years", "Years of Education",
ifelse(condition == "b_bfi_extra", "Extraversion",
ifelse(condition == "b_bfi_neuro", "Neuroticism",
ifelse(condition == "b_bfi_agree", "Agreeableness",
ifelse(condition == "b_bfi_consc", "Conscientiousness",
ifelse(condition == "b_bfi_open", "Openness",
ifelse(condition == "b_religiosity", "Religiosity",
condition)))))))))))))))),
group = ifelse(is.na(group), condition, group),
condition = factor(condition, levels = rev(c("Hormonal Contraception", "Age",
"500-1000 Euro", "1000-2000 Euro",
"2000-3000 Euro", ">3000 Euro", "do not tell",
"13-28 months", "29-52 months",
">52 months",
"Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))),
group = factor(group, levels = c("Contraception", "Age", "Income",
"Relationship Duration","Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.05))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.05, 0.05), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")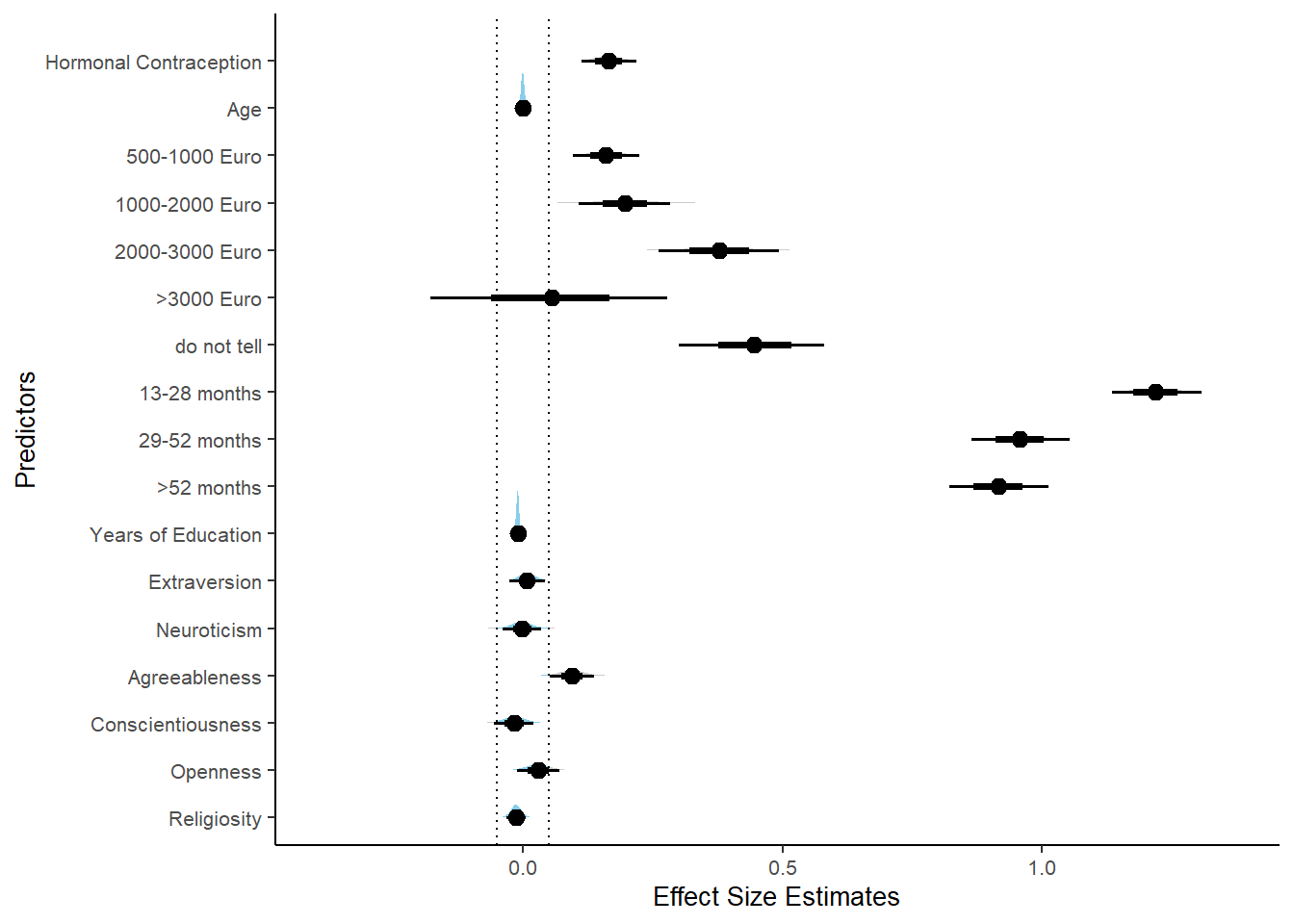
Masturabation Frequency
Model
m_hc_masfreq_controlled = brm(diary_masturbation_sum ~
offset(log(number_of_days)) +
contraception_hormonal +
age + net_income + relationship_duration_factor +
education_years +
bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open +
religiosity,
data = data, family = poisson(),
file = "m_hc_masfreq_controlled")Summary
## Family: poisson
## Links: mu = log
## Formula: diary_masturbation_sum ~ offset(log(number_of_days)) + contraception_hormonal + age + net_income + relationship_duration_factor + education_years + bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open + religiosity
## Data: data (Number of observations: 897)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept -1.67 0.19 -1.97 -1.36 1.00 3446
## contraception_hormonalyes -0.30 0.03 -0.34 -0.25 1.00 4457
## age -0.00 0.00 -0.01 0.00 1.00 4539
## net_incomeeuro_500_1000 0.18 0.03 0.13 0.24 1.00 3225
## net_incomeeuro_1000_2000 0.21 0.05 0.14 0.29 1.00 3055
## net_incomeeuro_2000_3000 0.05 0.07 -0.07 0.16 1.00 3731
## net_incomeeuro_gt_3000 -0.18 0.14 -0.42 0.05 1.00 4737
## net_incomedont_tell -0.22 0.10 -0.39 -0.05 1.00 4168
## relationship_duration_factorPartnered_upto12months -0.20 0.04 -0.26 -0.14 1.00 3456
## relationship_duration_factorPartnered_upto28months -0.23 0.04 -0.30 -0.17 1.00 3757
## relationship_duration_factorPartnered_upto52months -0.30 0.04 -0.36 -0.23 1.00 2965
## relationship_duration_factorPartnered_morethan52months -0.44 0.04 -0.51 -0.36 1.00 3239
## education_years 0.00 0.00 -0.00 0.01 1.00 6532
## bfi_extra -0.01 0.02 -0.04 0.02 1.00 3812
## bfi_neuro -0.01 0.02 -0.04 0.02 1.00 3190
## bfi_agree 0.00 0.02 -0.03 0.04 1.00 3875
## bfi_consc -0.21 0.02 -0.24 -0.17 1.00 4267
## bfi_open 0.20 0.02 0.17 0.24 1.00 4291
## religiosity -0.06 0.01 -0.07 -0.04 1.00 4998
## Tail_ESS
## Intercept 2984
## contraception_hormonalyes 3346
## age 3623
## net_incomeeuro_500_1000 3116
## net_incomeeuro_1000_2000 3063
## net_incomeeuro_2000_3000 3058
## net_incomeeuro_gt_3000 3281
## net_incomedont_tell 2963
## relationship_duration_factorPartnered_upto12months 2802
## relationship_duration_factorPartnered_upto28months 3112
## relationship_duration_factorPartnered_upto52months 2478
## relationship_duration_factorPartnered_morethan52months 3304
## education_years 3193
## bfi_extra 3226
## bfi_neuro 2866
## bfi_agree 2797
## bfi_consc 3059
## bfi_open 2873
## religiosity 3165
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_hc_masfreq_controlled, range = c(-0.05, 0.05), ci = 0.90,
parameters = "contraception"))## Picking joint bandwidth of 0.00384## Warning: Removed 399 rows containing non-finite values (stat_density_ridges).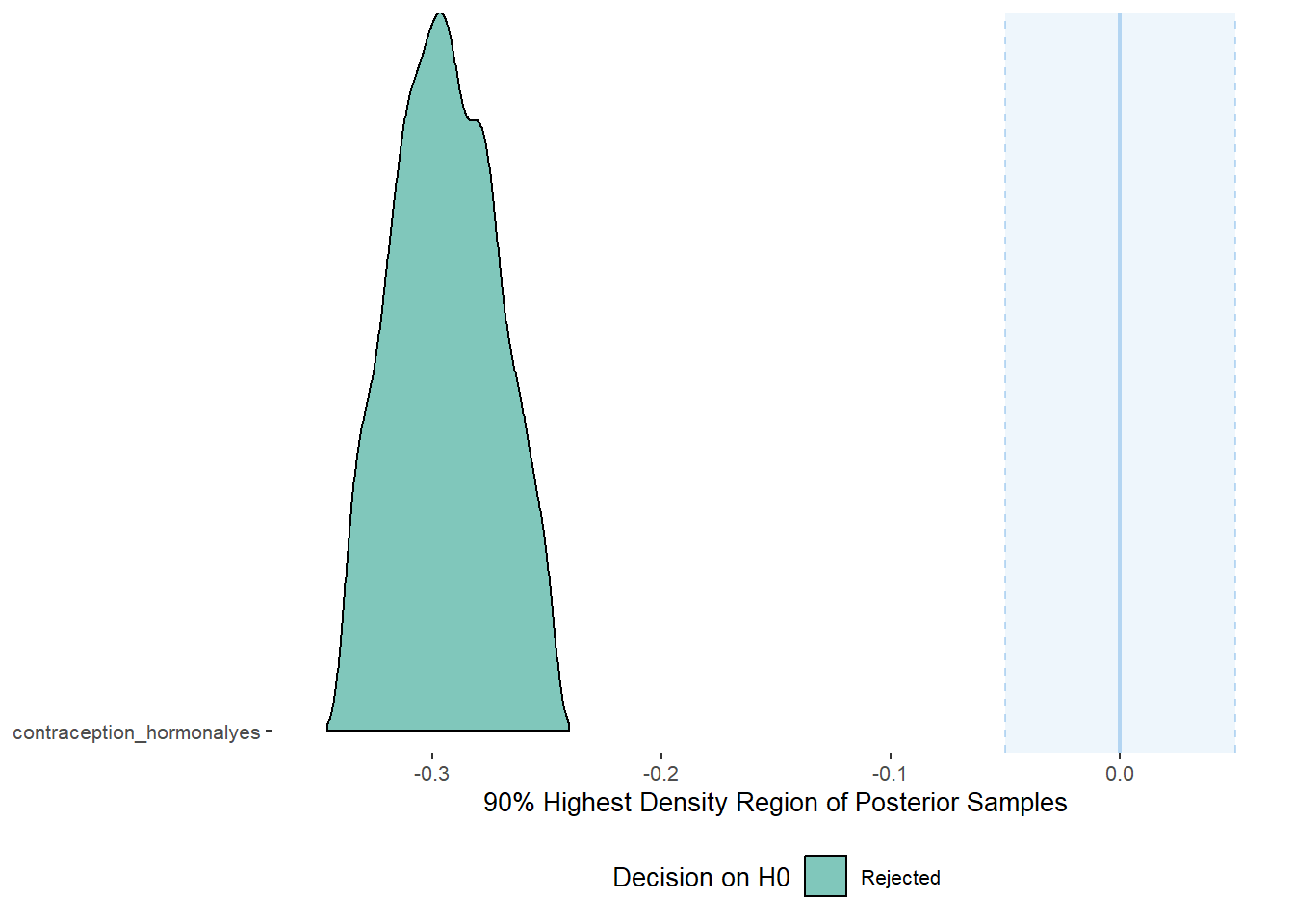
equivalence_test(m_hc_masfreq_controlled, range = c(-0.05, 0.05), ci = 0.90,
parameters = "contraception")## # A tibble: 1 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.05 0.05 0 Rejected -0.338 -0.248 fixed conditio~Plots
conditional_effects(m_hc_masfreq_controlled,
effects = "contraception_hormonal",
conditions = data.frame(number_of_days = 1))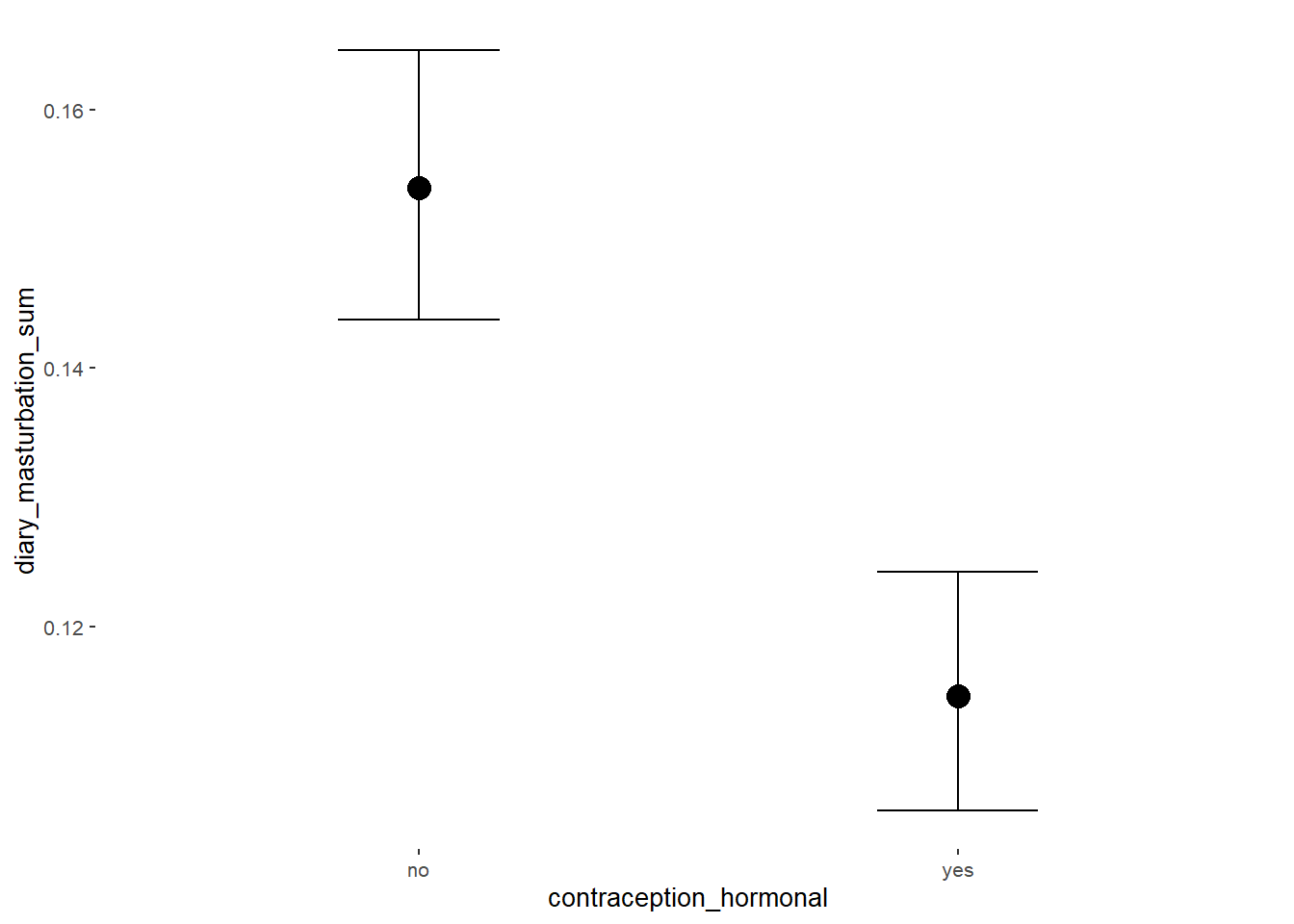
Forest Plot for Effect Sizes
m_hc_masfreq_controlled %>%
spread_draws(b_contraception_hormonalyes,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity) %>%
pivot_longer(cols = c(b_contraception_hormonalyes,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_relationship_duration_factor",
"Relationship Duration",
ifelse(condition %contains% "b_net_income",
"Income",
NA)),
group = ifelse(condition %contains% "b_contraception_hormonalyes",
"Contraception", group),
condition = ifelse(condition %contains% "b_contraception_hormonalyes",
"Hormonal Contraception", condition),
condition = ifelse(condition == "b_age", "Age",
ifelse(condition == "b_net_incomeeuro_500_1000", "500-1000 Euro",
ifelse(condition == "b_net_incomeeuro_1000_2000", "1000-2000 Euro",
ifelse(condition == "b_net_incomeeuro_2000_3000", "2000-3000 Euro",
ifelse(condition == "b_net_incomeeuro_gt_3000", ">3000 Euro",
ifelse(condition == "b_net_incomedont_tell", "do not tell",
ifelse(condition == "b_relationship_duration_factorPartnered_upto28months",
"13-28 months",
ifelse(condition == "b_relationship_duration_factorPartnered_upto52months",
"29-52 months",
ifelse(condition == "b_relationship_duration_factorPartnered_morethan52months",
">52 months",
ifelse(condition == "b_education_years", "Years of Education",
ifelse(condition == "b_bfi_extra", "Extraversion",
ifelse(condition == "b_bfi_neuro", "Neuroticism",
ifelse(condition == "b_bfi_agree", "Agreeableness",
ifelse(condition == "b_bfi_consc", "Conscientiousness",
ifelse(condition == "b_bfi_open", "Openness",
ifelse(condition == "b_religiosity", "Religiosity",
condition)))))))))))))))),
group = ifelse(is.na(group), condition, group),
condition = factor(condition, levels = rev(c("Hormonal Contraception", "Age",
"500-1000 Euro", "1000-2000 Euro",
"2000-3000 Euro", ">3000 Euro", "do not tell",
"13-28 months", "29-52 months",
">52 months",
"Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))),
group = factor(group, levels = c("Contraception", "Age", "Income",
"Relationship Duration","Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.05))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.05, 0.05), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")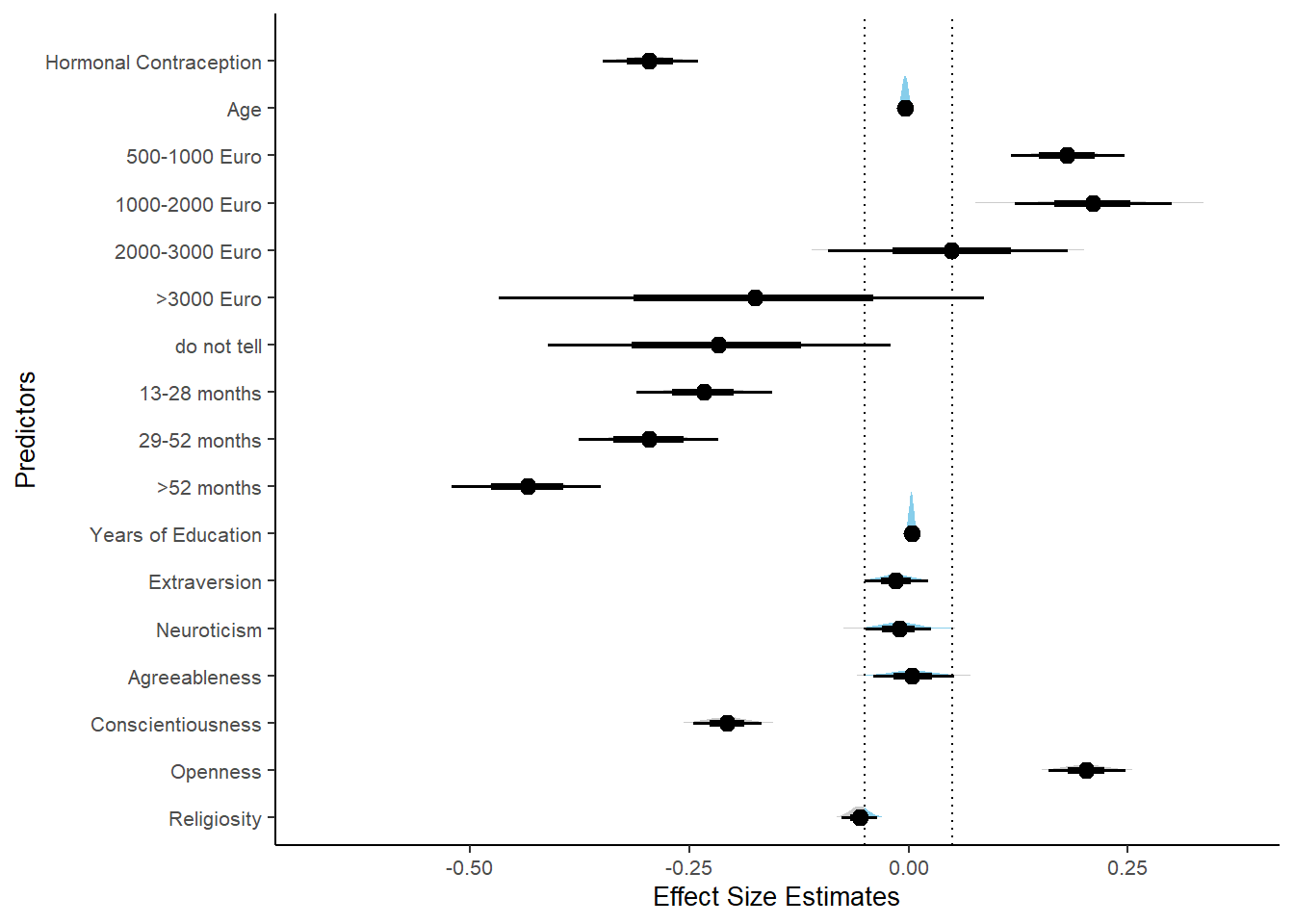
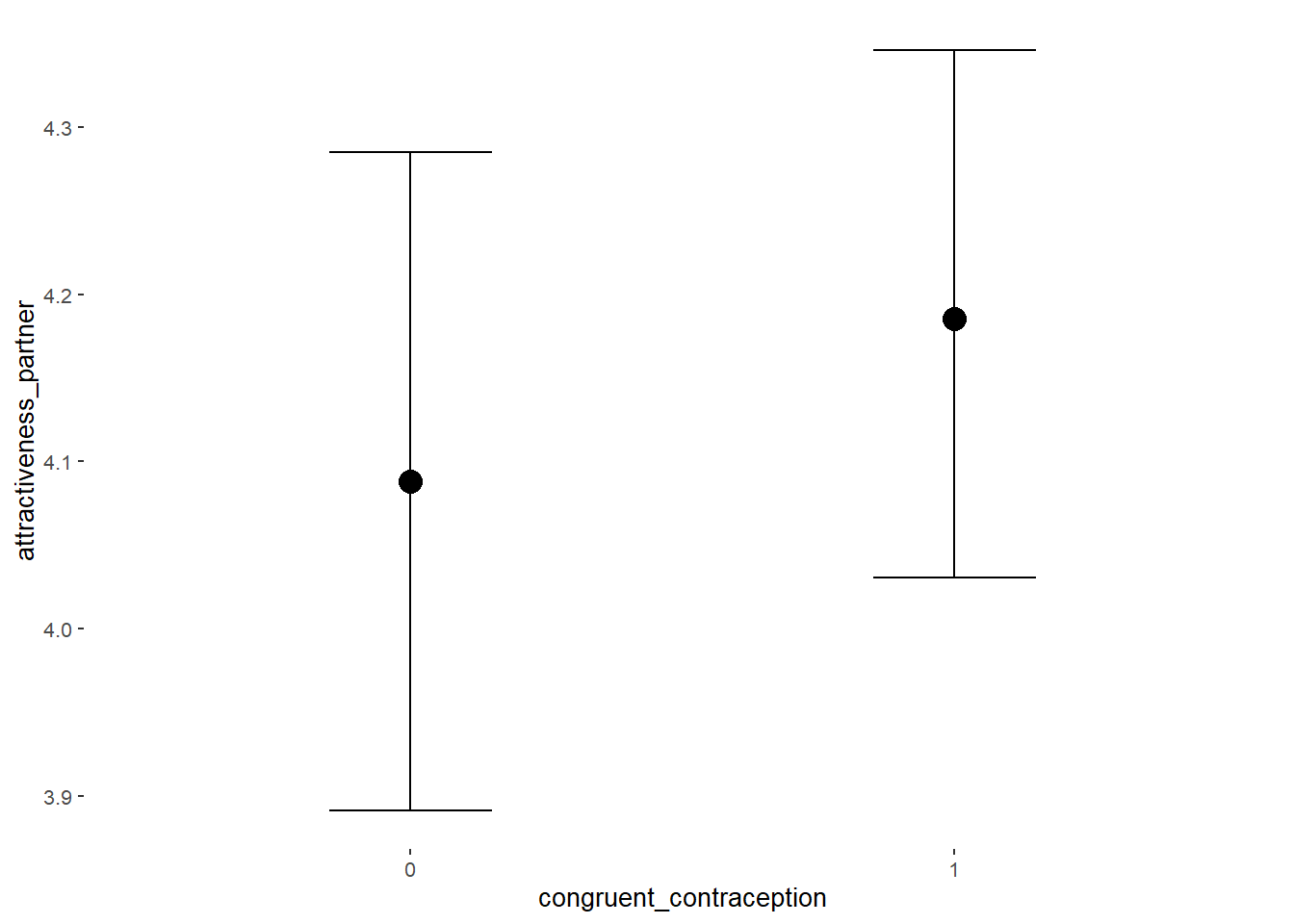
Effects of (In)Congruenct HC Use
Attractiveness of Partner
Model
Summary
## Family: gaussian
## Links: mu = identity; sigma = identity
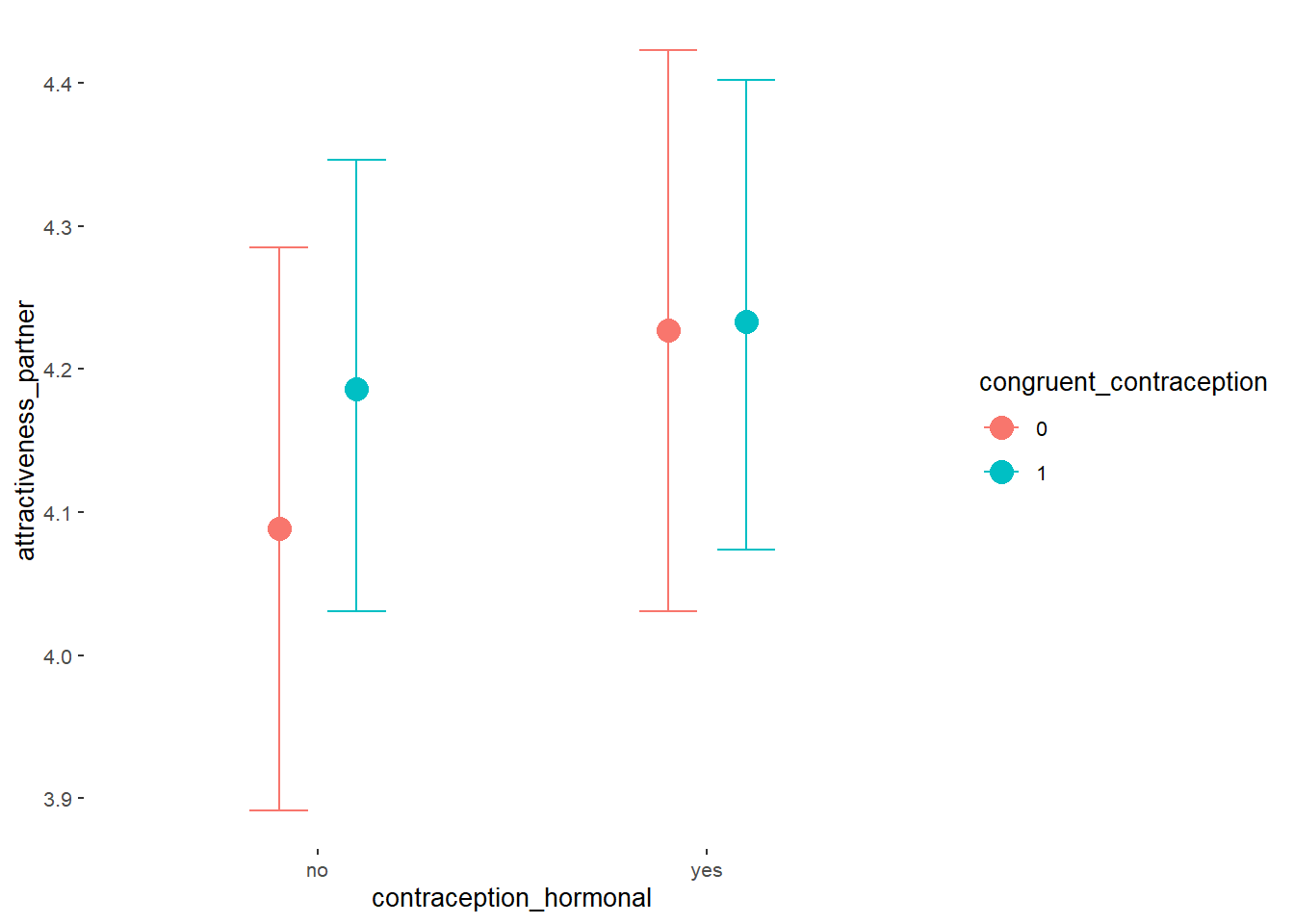
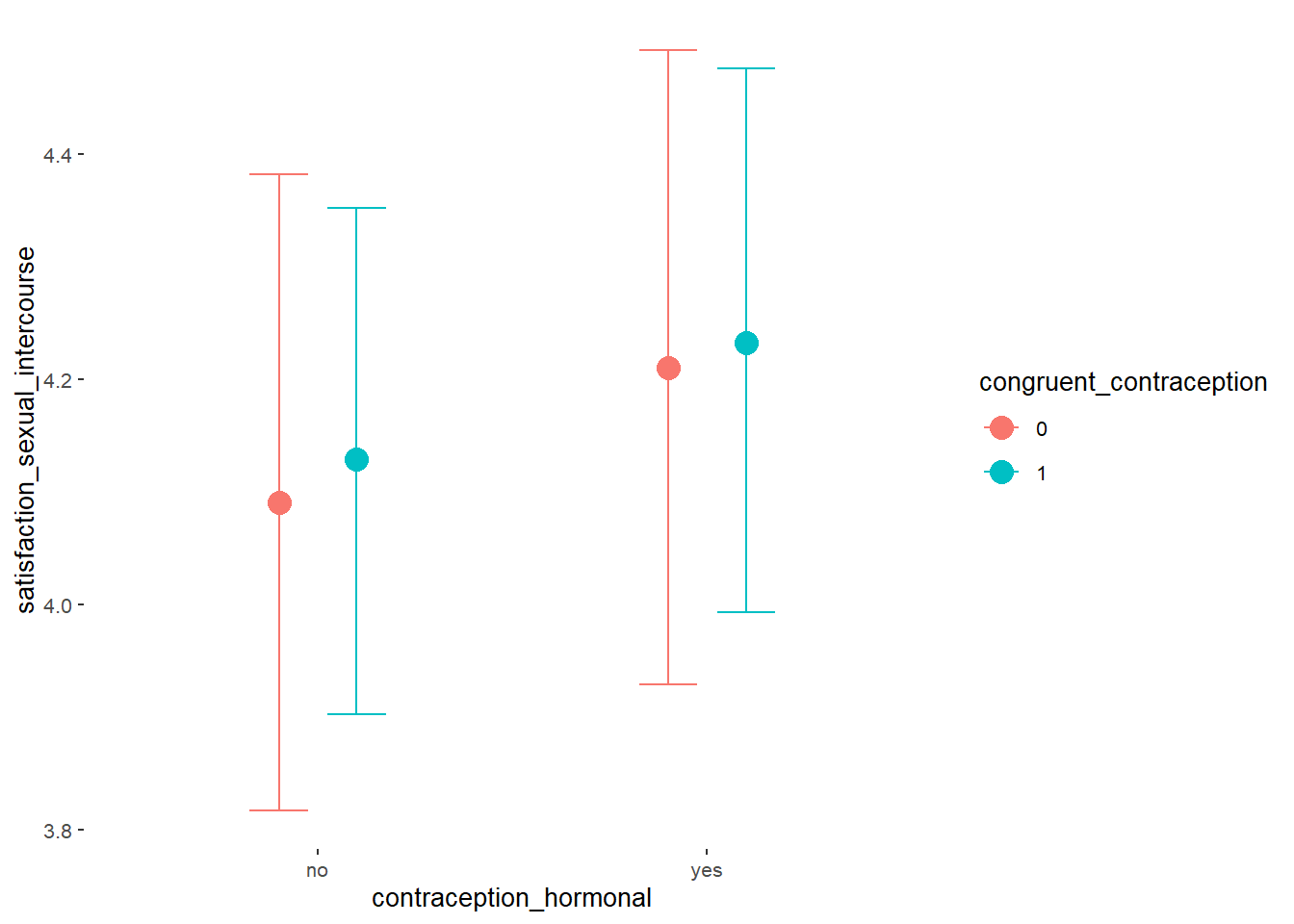
## Formula: attractiveness_partner ~ contraception_hormonal * congruent_contraception
## Data: data (Number of observations: 774)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
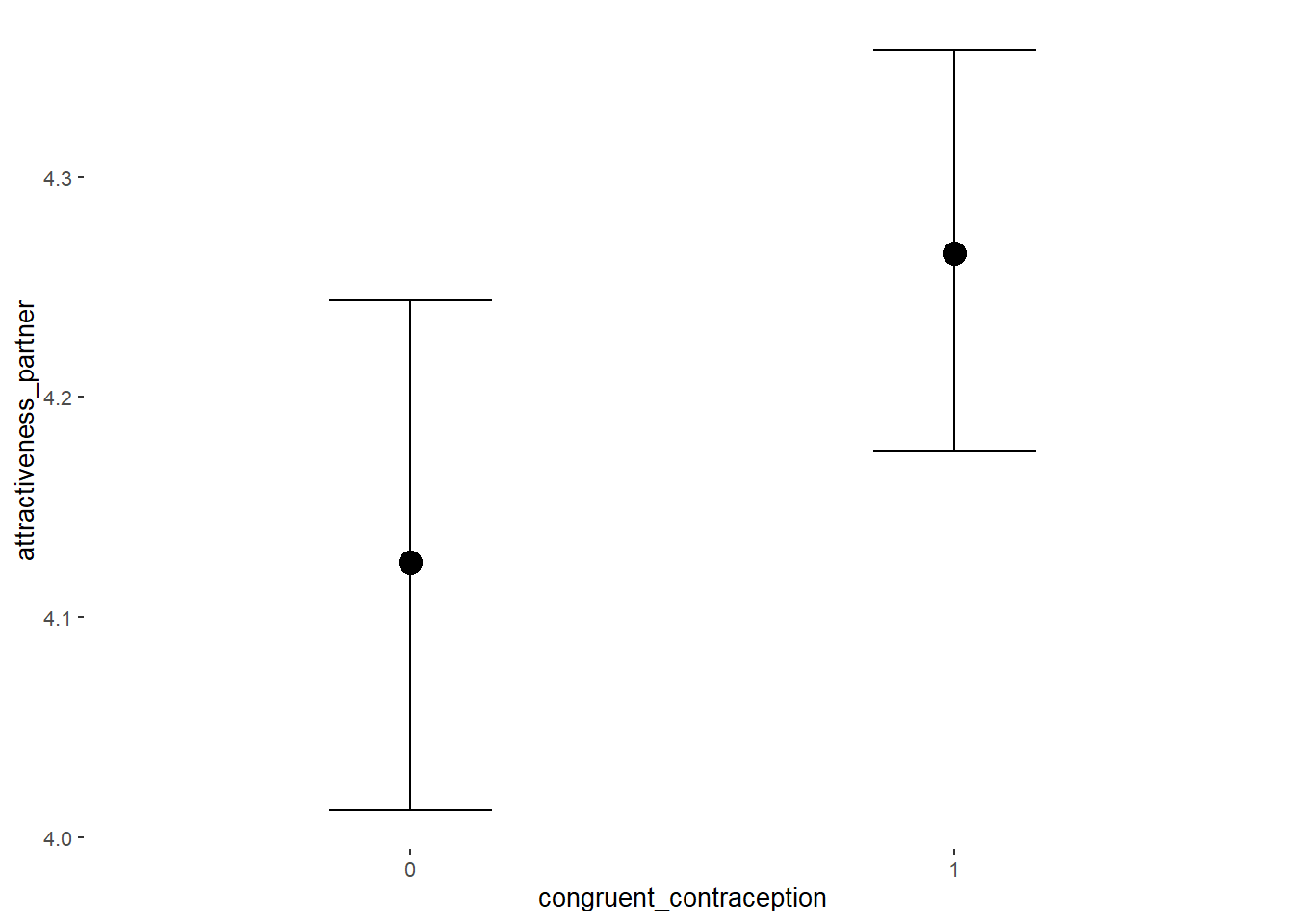
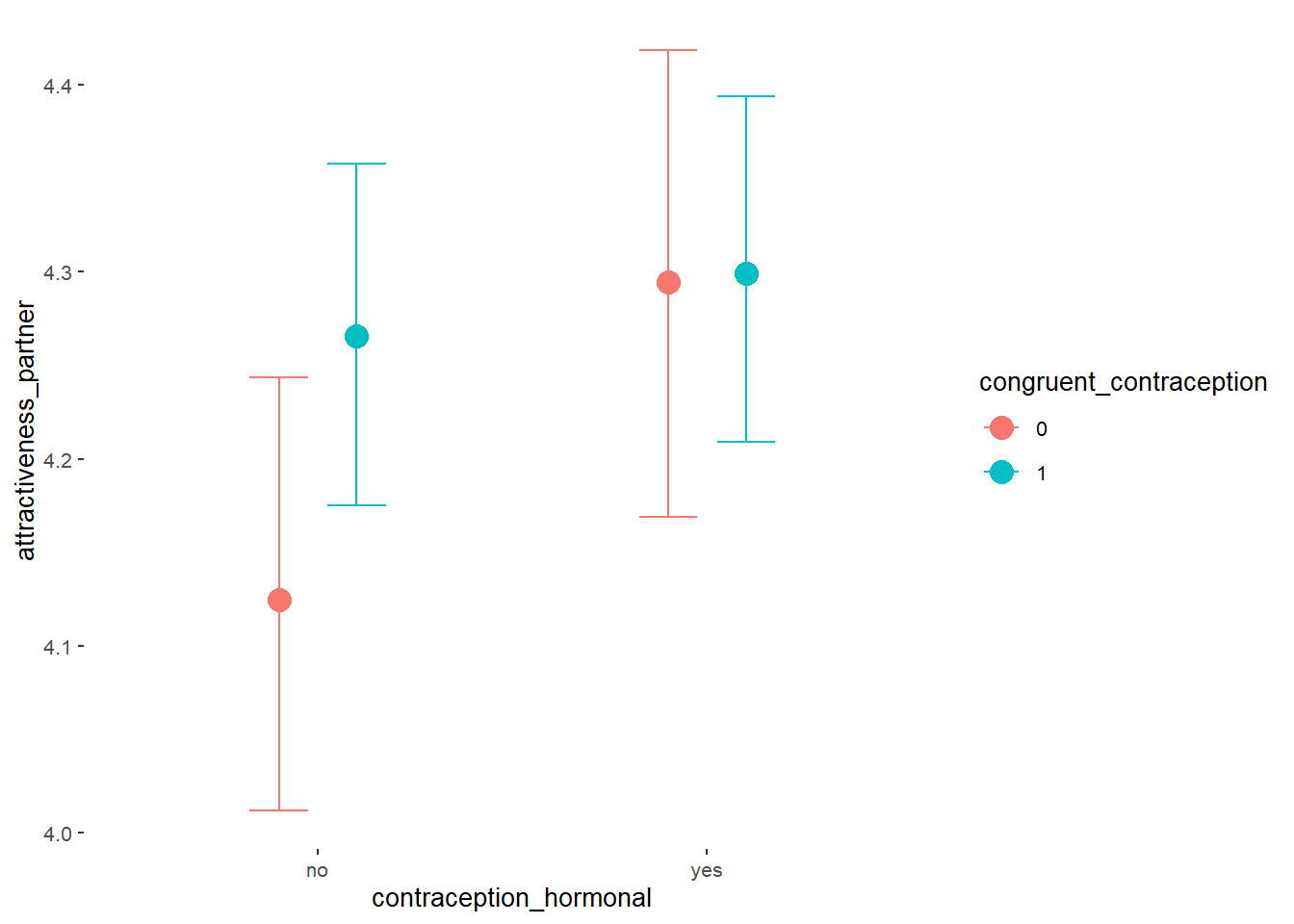
## Intercept 4.13 0.06 4.03 4.23 1.00 2377
## contraception_hormonalyes 0.17 0.09 0.02 0.31 1.00 1982
## congruent_contraception1 0.14 0.08 0.01 0.27 1.00 2228
## contraception_hormonalyes:congruent_contraception1 -0.13 0.11 -0.31 0.04 1.00 1877
## Tail_ESS
## Intercept 2715
## contraception_hormonalyes 2296
## congruent_contraception1 2605
## contraception_hormonalyes:congruent_contraception1 2380
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.74 0.02 0.71 0.77 1.00 2990 2773
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_congruency_atrr, range = c(-0.07, 0.07), ci = 0.90,
parameters = "contraception"))## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.79). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## Picking joint bandwidth of 0.0125## Warning: Removed 1197 rows containing non-finite values (stat_density_ridges).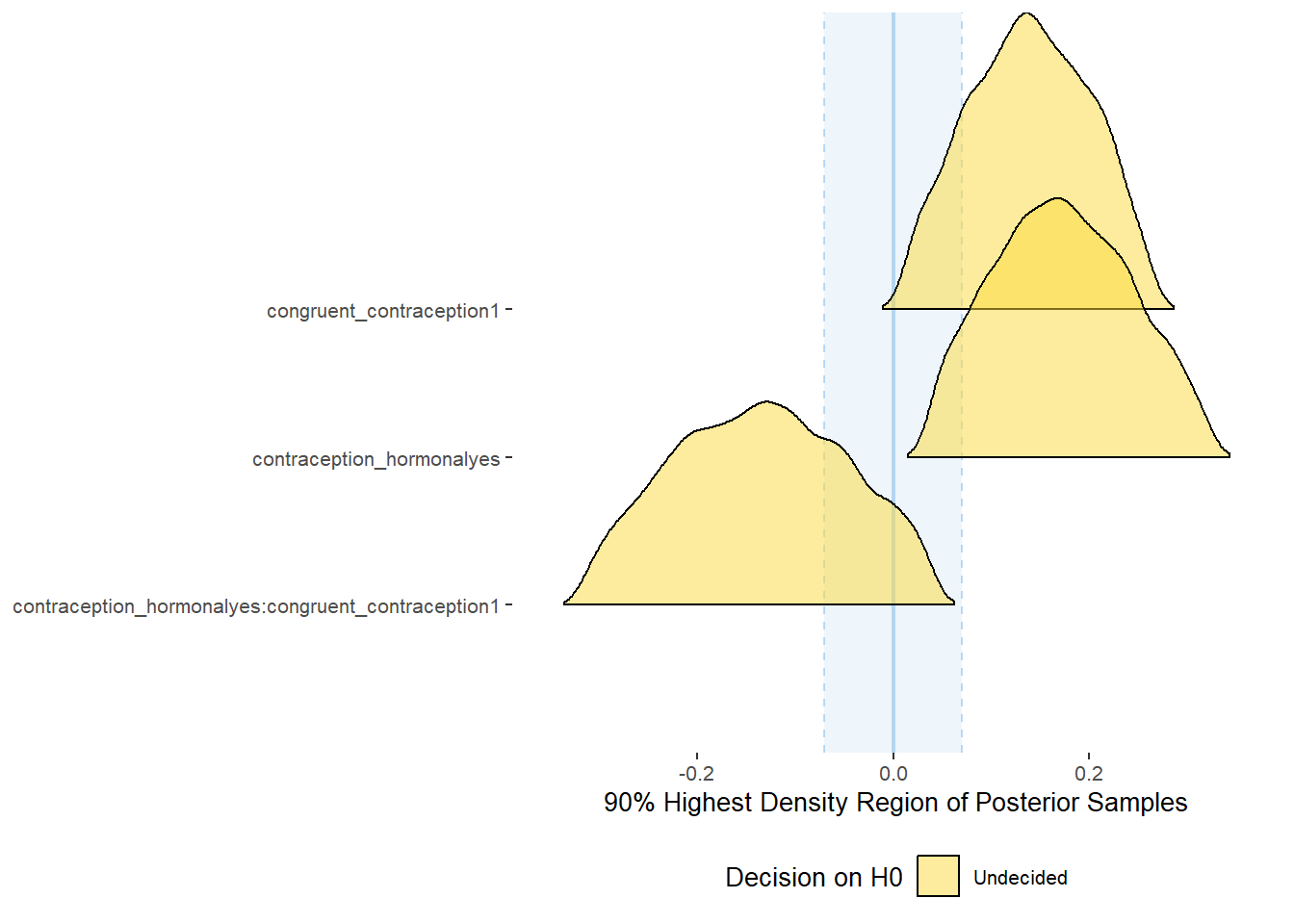
equivalence_test(m_congruency_atrr, range = c(-0.07, 0.07), ci = 0.90,
parameters = "contraception")## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.79). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## # A tibble: 3 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.07 0.07 0.0711 Undecided 0.0377 0.322 fixed conditio~
## 2 b_congruent_co~ 0.9 -0.07 0.07 0.150 Undecided 0.0128 0.263 fixed conditio~
## 3 b_contraceptio~ 0.9 -0.07 0.07 0.256 Undecided -0.315 0.0417 fixed conditio~Forest Plot for Effect Sizes
m_congruency_atrr %>%
spread_draws(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`) %>%
pivot_longer(cols = c(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "ontraception",
"Contraception", NA),
condition = ifelse(condition == "b_contraception_hormonalyes",
"Hormonal Contraception",
ifelse(condition == "b_congruent_contraception1",
"Congruent Contraception",
ifelse(condition == "b_contraception_hormonalyes:congruent_contraception1",
"Interaction Hormonal Contracpetion and Congruent Contraception",
condition))),
condition = factor(condition, levels = rev(c("Hormonal Contraception",
"Congruent Contraception",
"Interaction Hormonal Contracpetion and Congruent Contraception")))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.07))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.07, 0.07), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors") +
xlim (-0.6, 0.6)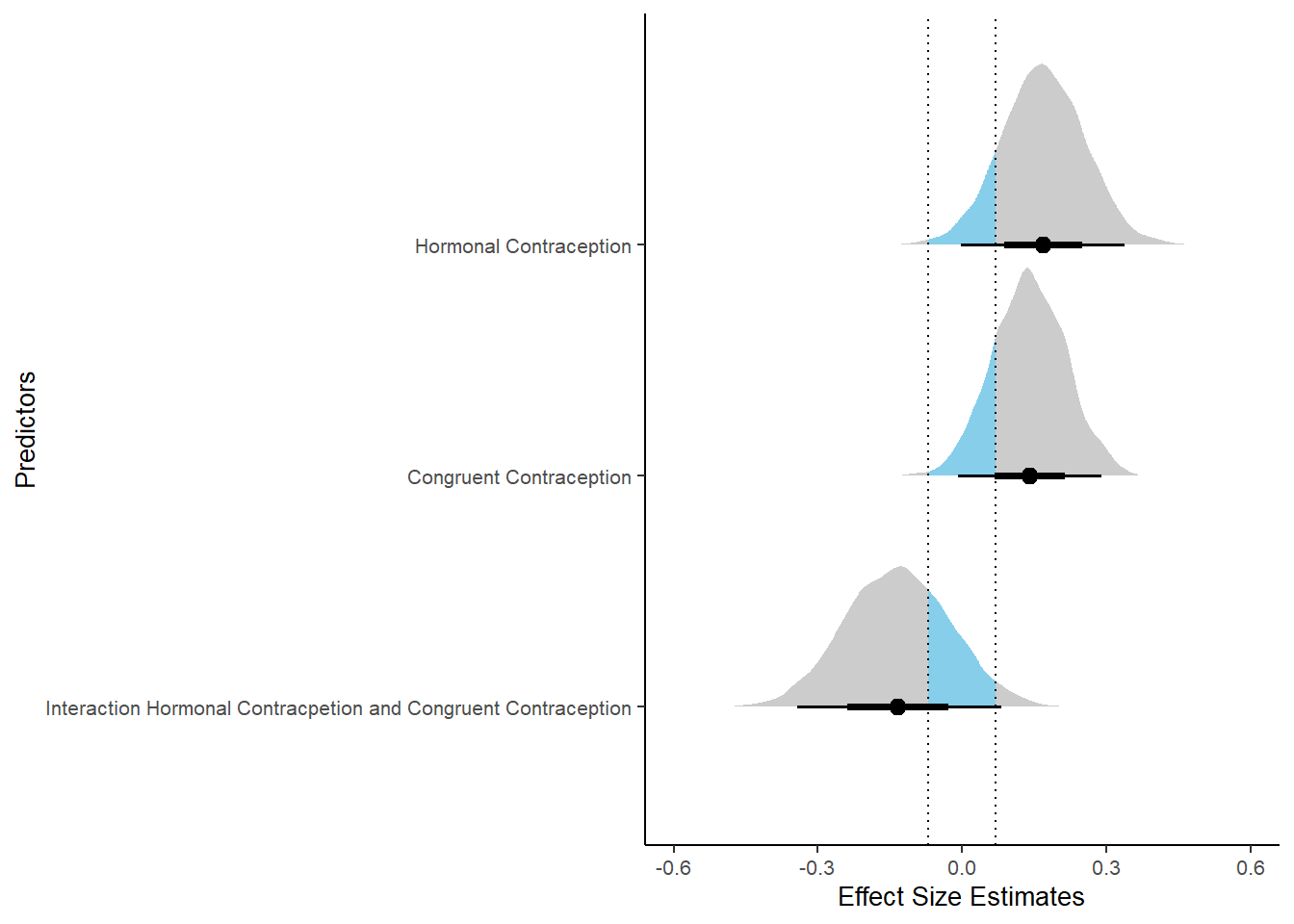
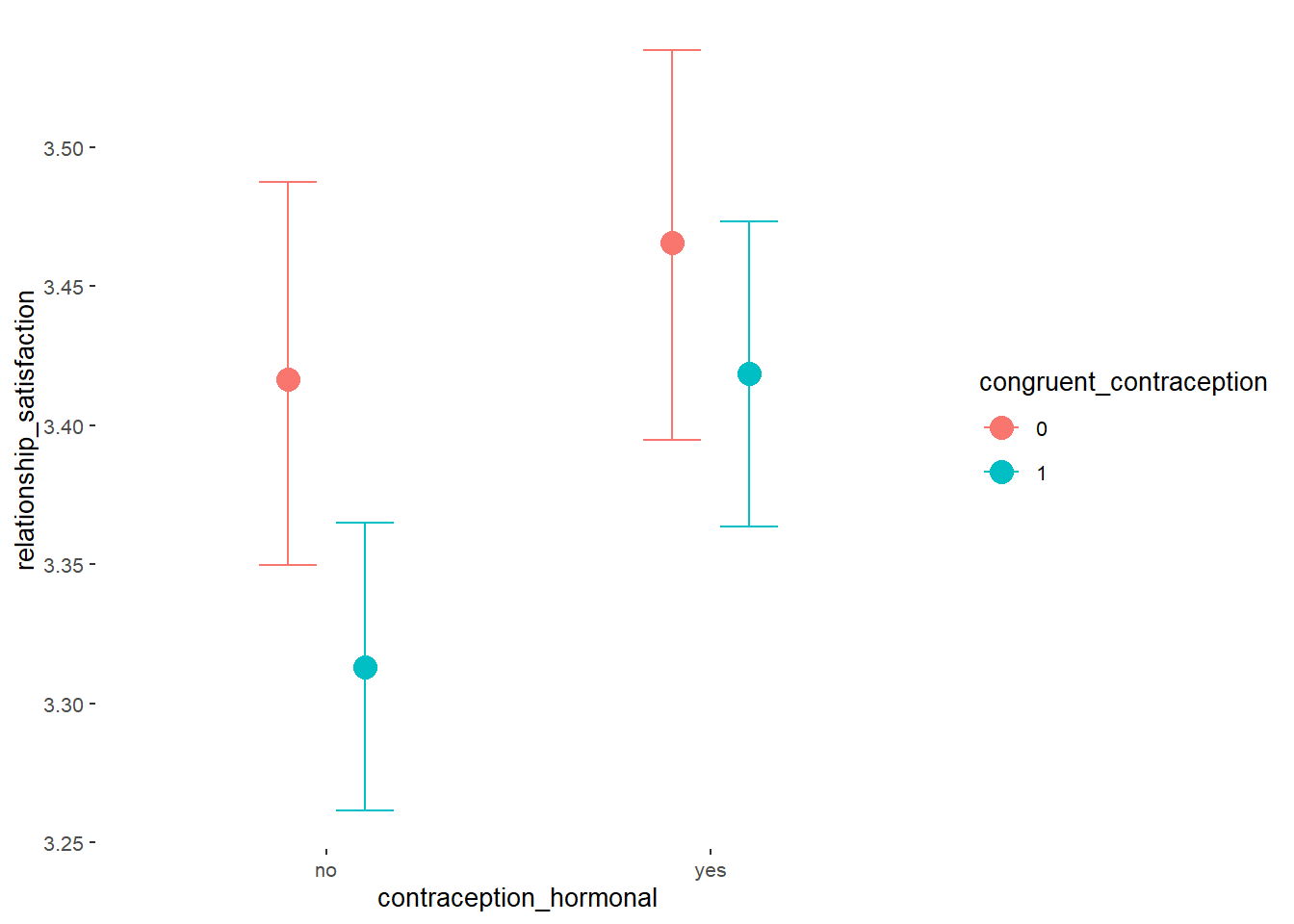
Relationship Satisfaction
Model
Summary
## Family: gaussian
## Links: mu = identity; sigma = identity
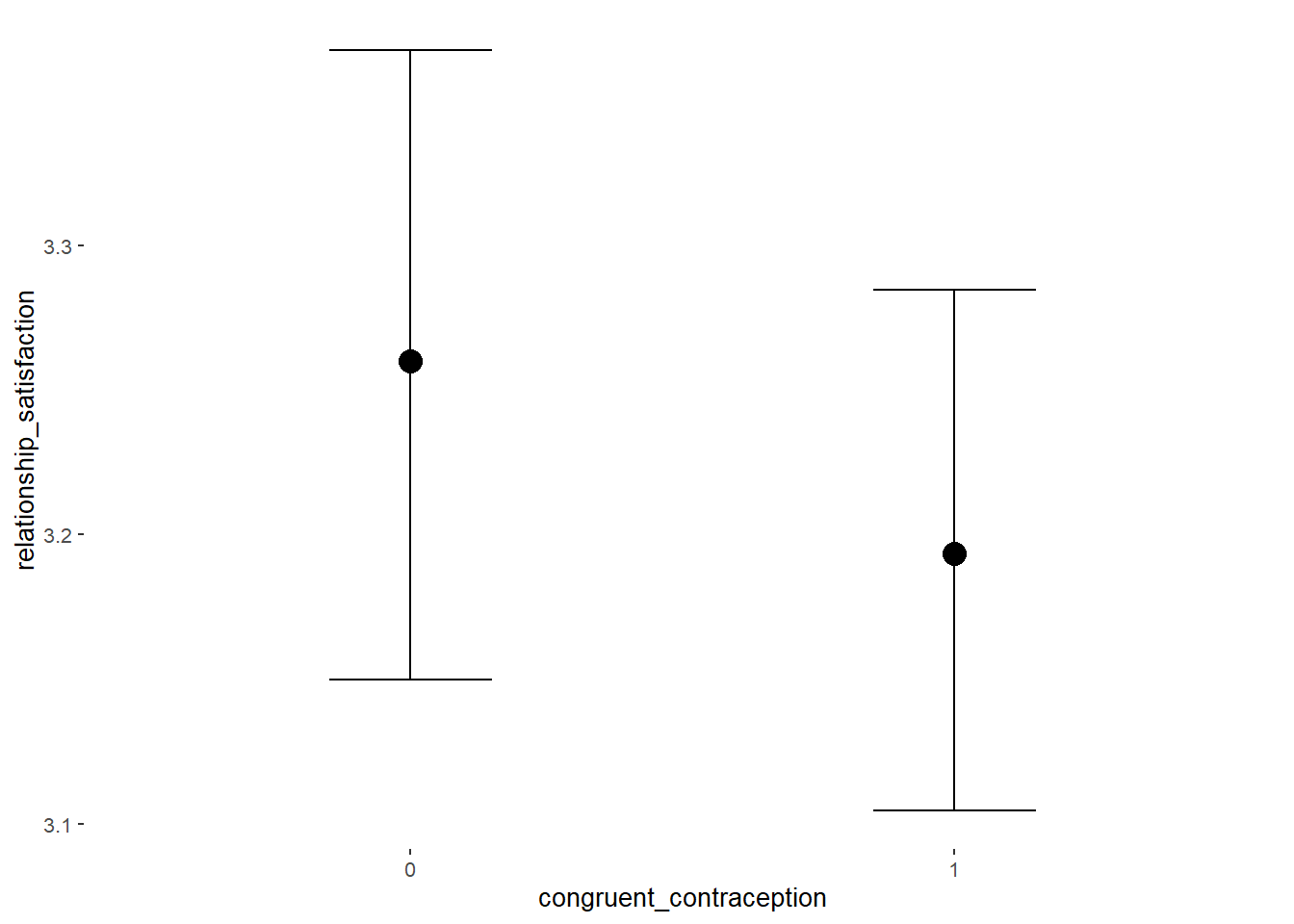
## Formula: relationship_satisfaction ~ contraception_hormonal * congruent_contraception
## Data: data (Number of observations: 774)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept 3.42 0.04 3.36 3.47 1.00 2107
## contraception_hormonalyes 0.05 0.05 -0.03 0.13 1.00 1811
## congruent_contraception1 -0.10 0.04 -0.18 -0.03 1.00 1922
## contraception_hormonalyes:congruent_contraception1 0.06 0.06 -0.05 0.16 1.00 1720
## Tail_ESS
## Intercept 2335
## contraception_hormonalyes 2605
## congruent_contraception1 2314
## contraception_hormonalyes:congruent_contraception1 2381
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.42 0.01 0.41 0.44 1.01 3232 2151
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_congruency_relsat, range = c(-0.04, 0.04), ci = 0.90,
parameters = "contraception"))## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.79), b_contraception_hormonalyes:congruent_contraception1 and b_congruent_contraception1 (r = 0.71). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## Picking joint bandwidth of 0.00737## Warning: Removed 1197 rows containing non-finite values (stat_density_ridges).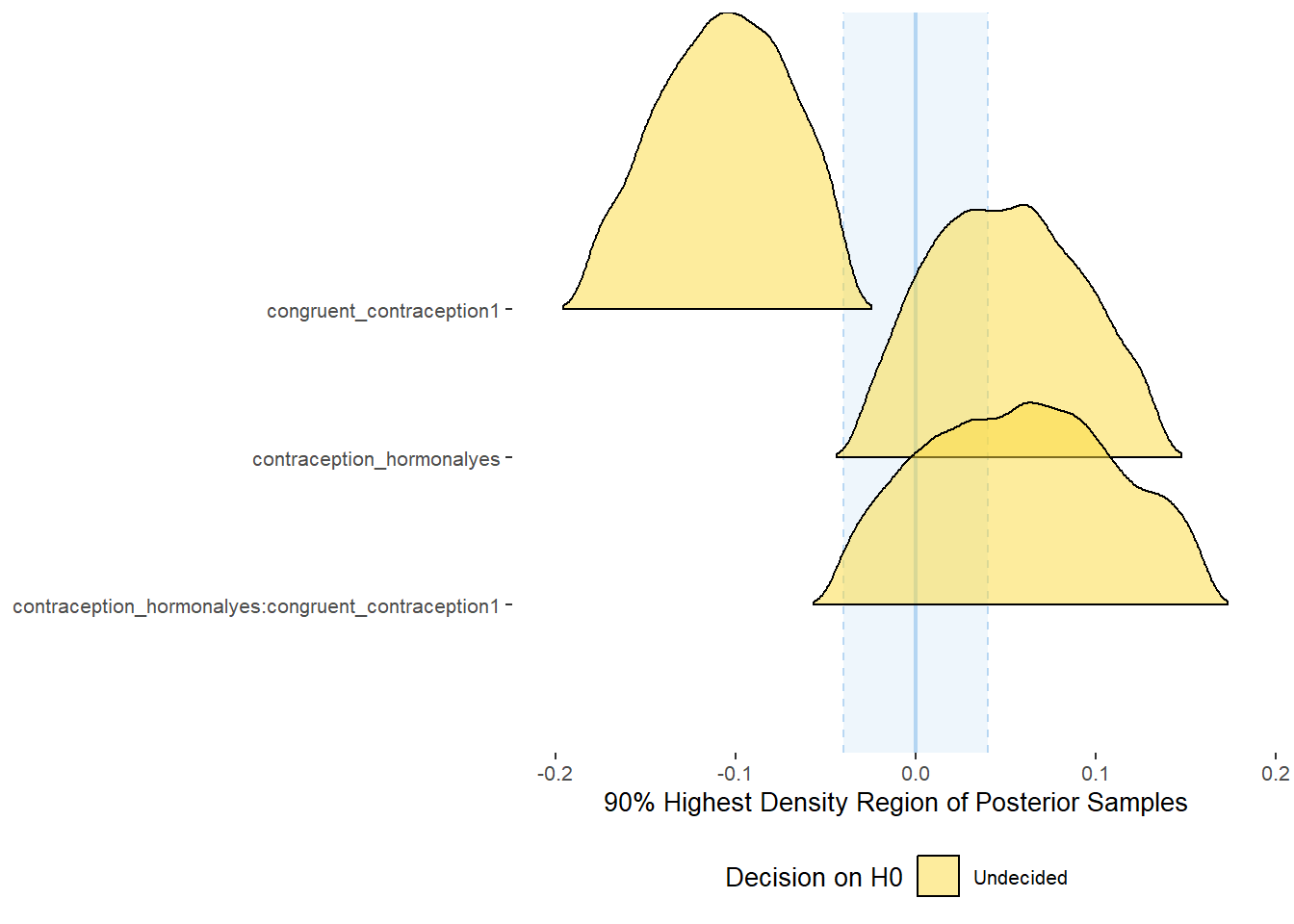
equivalence_test(m_congruency_relsat, range = c(-0.04, 0.04), ci = 0.90,
parameters = "contraception")## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.79), b_contraception_hormonalyes:congruent_contraception1 and b_congruent_contraception1 (r = 0.71). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## # A tibble: 3 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.04 0.04 0.425 Undecided -0.0300 0.135 fixed conditio~
## 2 b_congruent_co~ 0.9 -0.04 0.04 0.00555 Undecided -0.183 -0.0386 fixed conditio~
## 3 b_contraceptio~ 0.9 -0.04 0.04 0.377 Undecided -0.0439 0.161 fixed conditio~Forest Plot for Effect Sizes
m_congruency_relsat %>%
spread_draws(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`) %>%
pivot_longer(cols = c(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "ontraception",
"Contraception", NA),
condition = ifelse(condition == "b_contraception_hormonalyes",
"Hormonal Contraception",
ifelse(condition == "b_congruent_contraception1",
"Congruent Contraception",
ifelse(condition == "b_contraception_hormonalyes:congruent_contraception1",
"Interaction Hormonal Contracpetion and Congruent Contraception",
condition))),
condition = factor(condition, levels = rev(c("Hormonal Contraception",
"Congruent Contraception",
"Interaction Hormonal Contracpetion and Congruent Contraception")))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.04))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.04, 0.04), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors") +
xlim (-0.6, 0.6)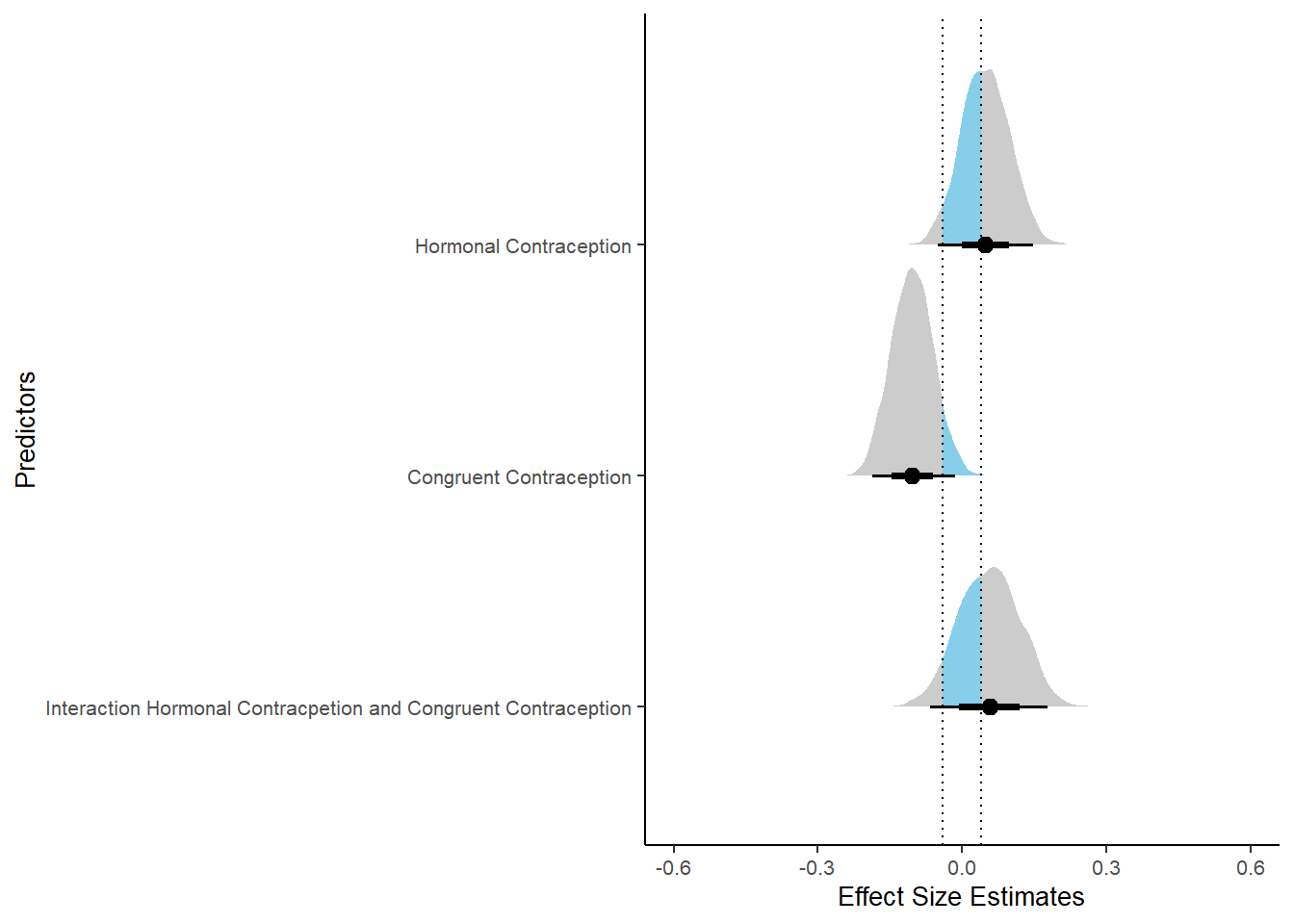
Sexual Satisfaction
Model
Summary
## Family: gaussian
## Links: mu = identity; sigma = identity
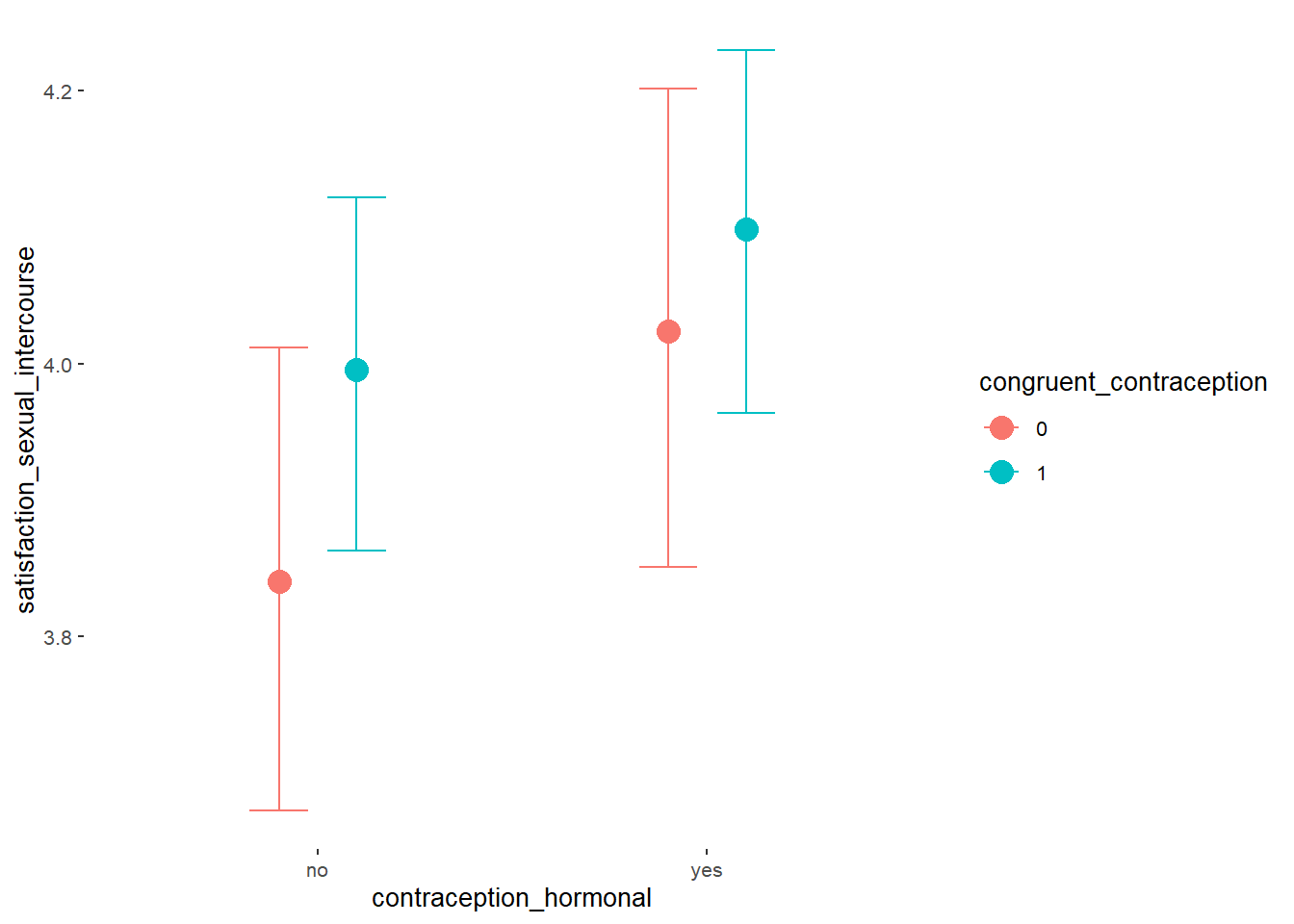
## Formula: satisfaction_sexual_intercourse ~ contraception_hormonal * congruent_contraception
## Data: data (Number of observations: 774)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept 3.84 0.09 3.70 3.98 1.00 1861
## contraception_hormonalyes 0.18 0.12 -0.02 0.39 1.00 1693
## congruent_contraception1 0.15 0.11 -0.03 0.33 1.00 1958
## contraception_hormonalyes:congruent_contraception1 -0.08 0.16 -0.33 0.19 1.00 1602
## Tail_ESS
## Intercept 2624
## contraception_hormonalyes 2227
## congruent_contraception1 2102
## contraception_hormonalyes:congruent_contraception1 2198
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 1.05 0.03 1.01 1.10 1.00 3579 2812
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_congruency_sexsat, range = c(-0.11, 0.11), ci = 0.90,
parameters = "contraception"))## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.8), b_contraception_hormonalyes:congruent_contraception1 and b_congruent_contraception1 (r = 0.7). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## Picking joint bandwidth of 0.018## Warning: Removed 1197 rows containing non-finite values (stat_density_ridges).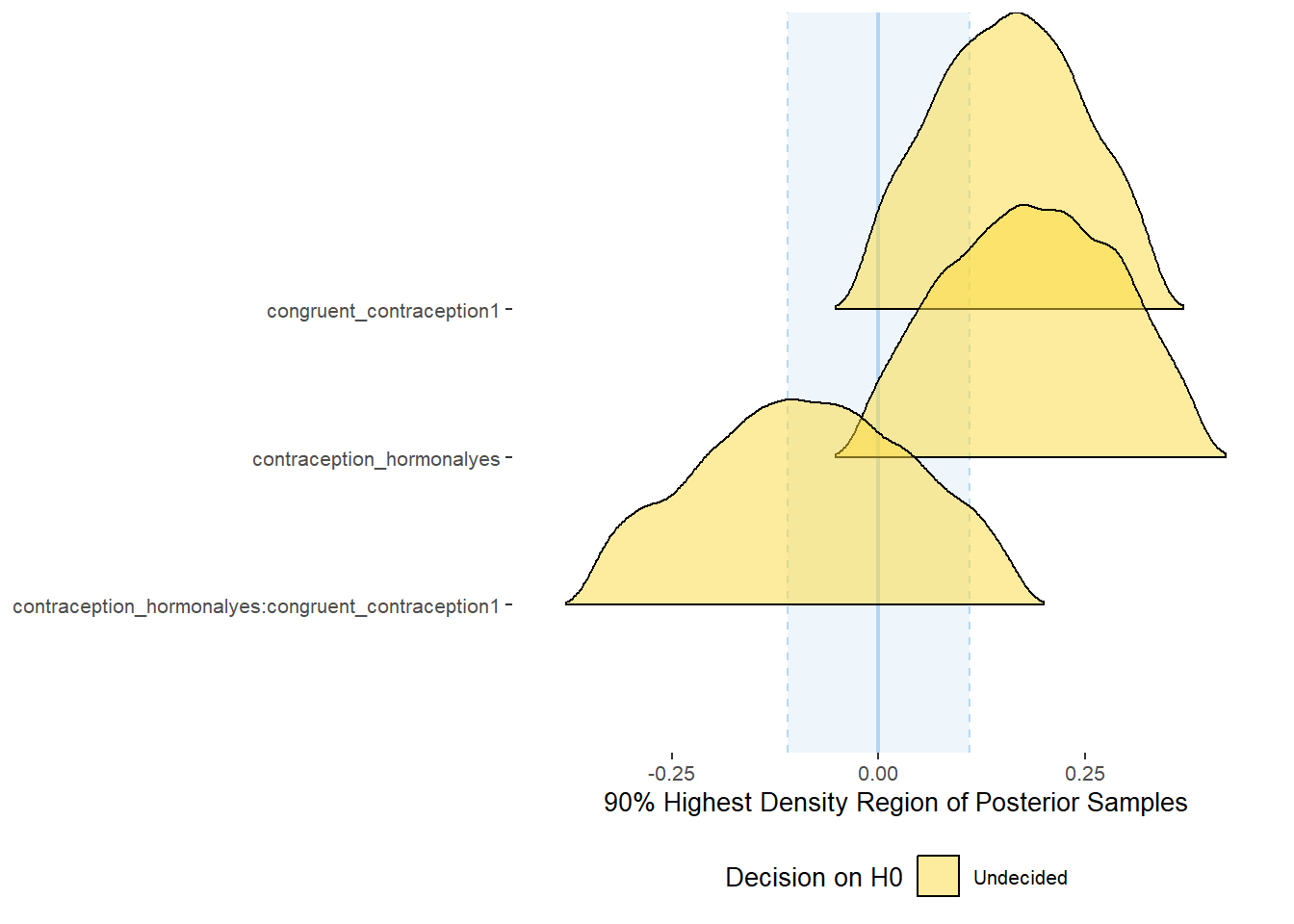
equivalence_test(m_congruency_sexsat, range = c(-0.11, 0.11), ci = 0.90,
parameters = "contraception")## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.8), b_contraception_hormonalyes:congruent_contraception1 and b_congruent_contraception1 (r = 0.7). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## # A tibble: 3 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.11 0.11 0.249 Undecided -0.0184 0.390 fixed conditio~
## 2 b_congruent_co~ 0.9 -0.11 0.11 0.313 Undecided -0.0187 0.335 fixed conditio~
## 3 b_contraceptio~ 0.9 -0.11 0.11 0.500 Undecided -0.347 0.171 fixed conditio~Forest Plot for Effect Sizes
m_congruency_sexsat %>%
spread_draws(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`) %>%
pivot_longer(cols = c(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "ontraception",
"Contraception", NA),
condition = ifelse(condition == "b_contraception_hormonalyes",
"Hormonal Contraception",
ifelse(condition == "b_congruent_contraception1",
"Congruent Contraception",
ifelse(condition == "b_contraception_hormonalyes:congruent_contraception1",
"Interaction Hormonal Contracpetion and Congruent Contraception",
condition))),
condition = factor(condition, levels = rev(c("Hormonal Contraception",
"Congruent Contraception",
"Interaction Hormonal Contracpetion and Congruent Contraception")))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.11))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.11, 0.11), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors") +
xlim (-0.6, 0.6)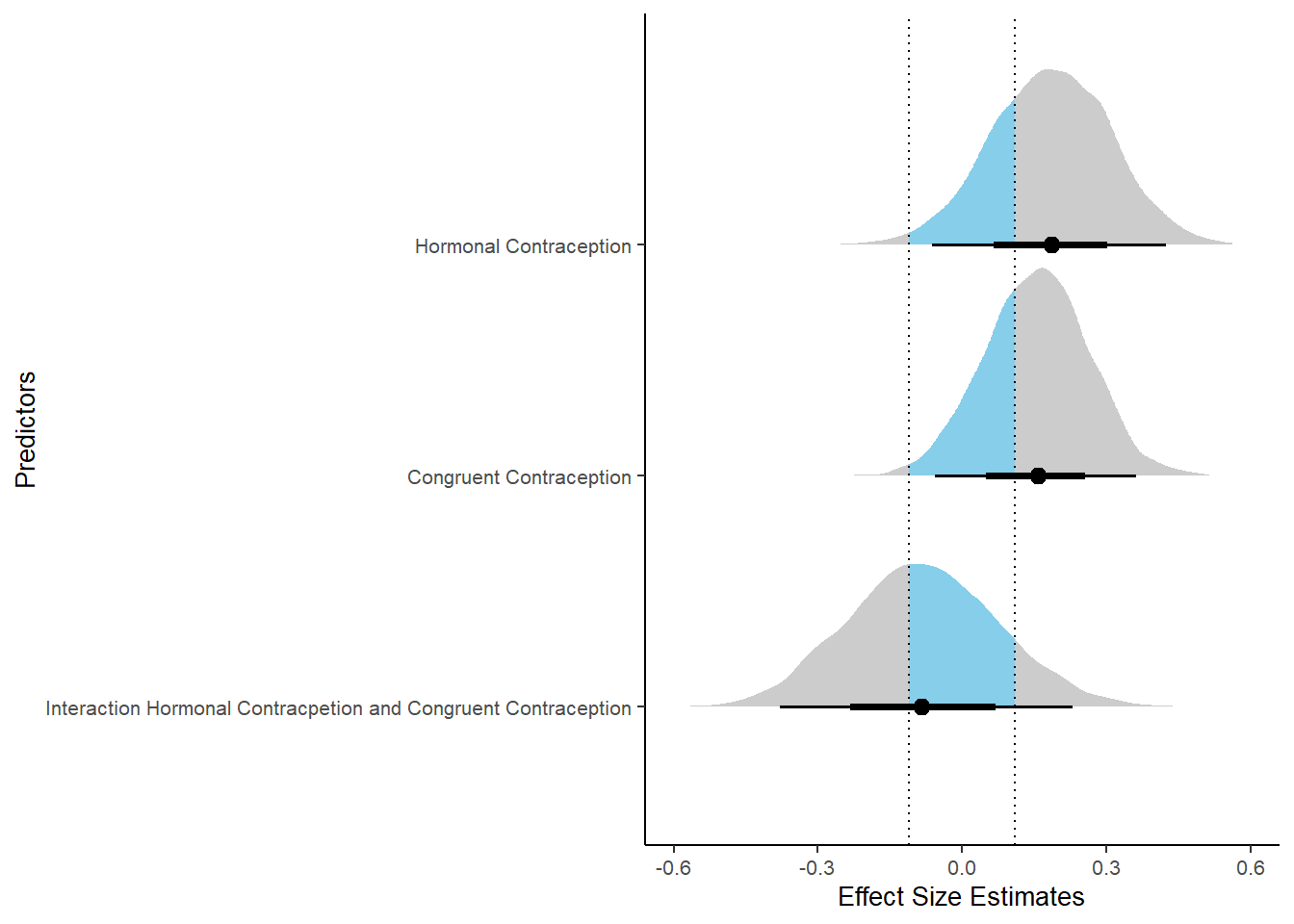
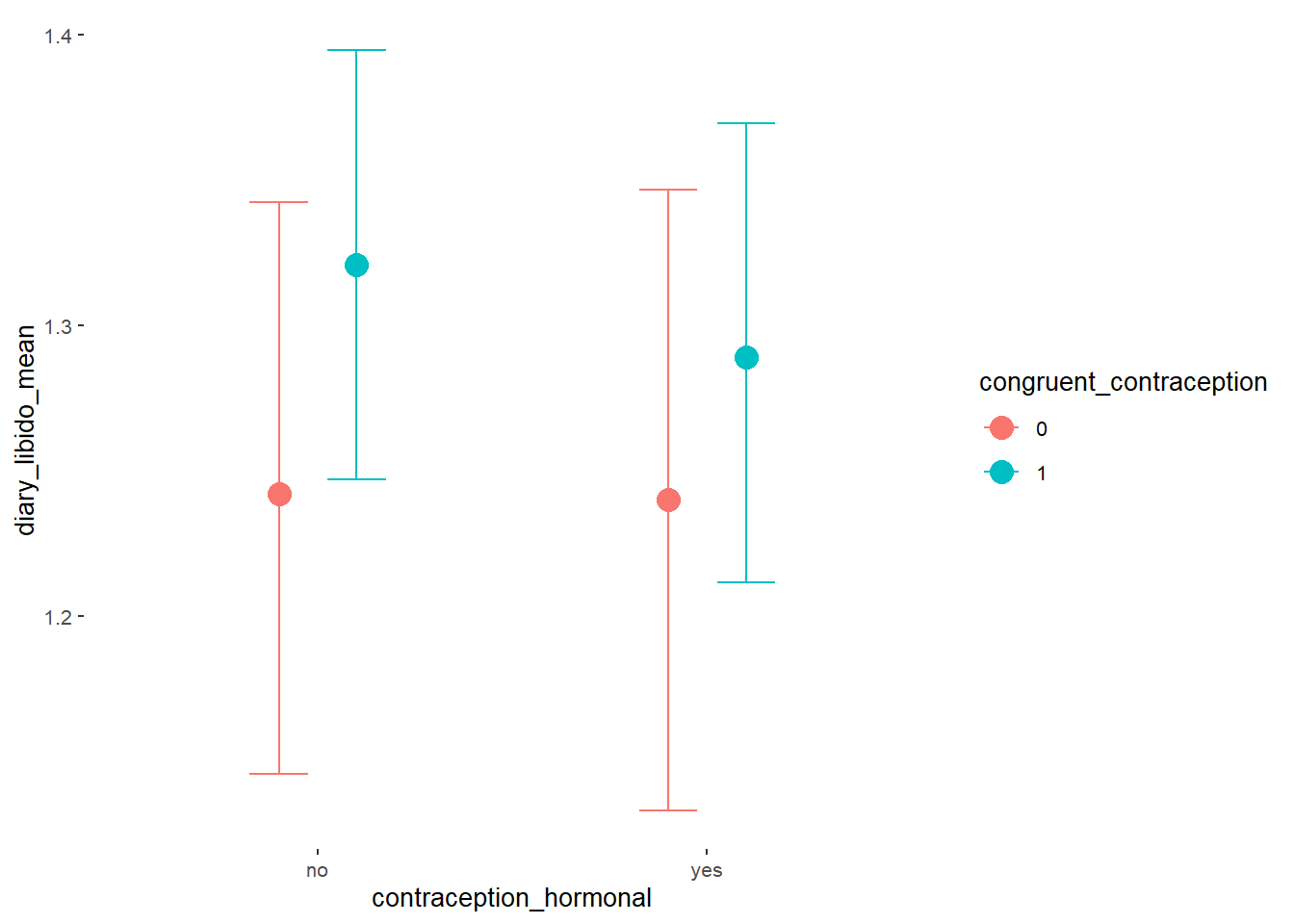
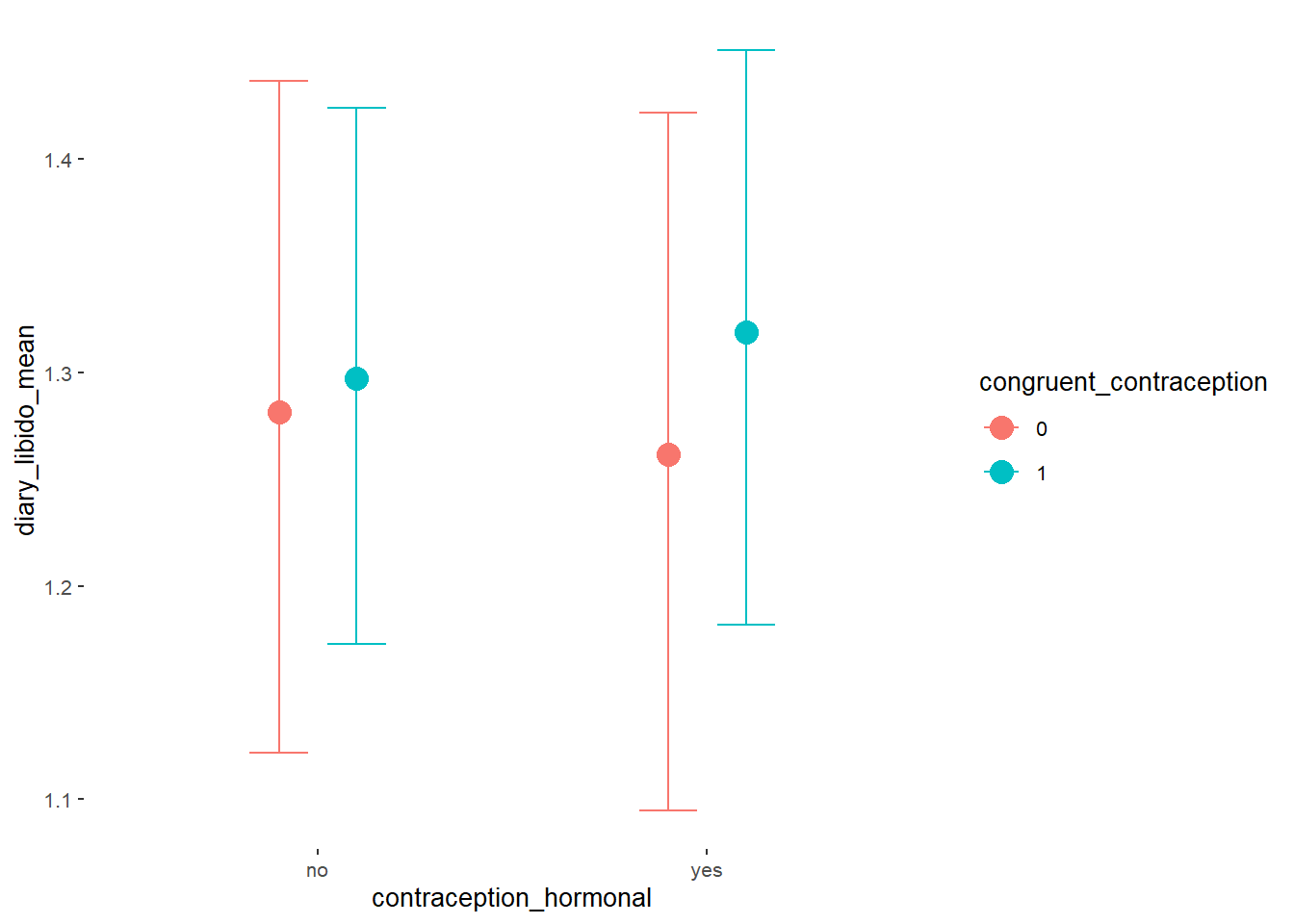
Libido
Model
Summary
## Family: gaussian
## Links: mu = identity; sigma = identity
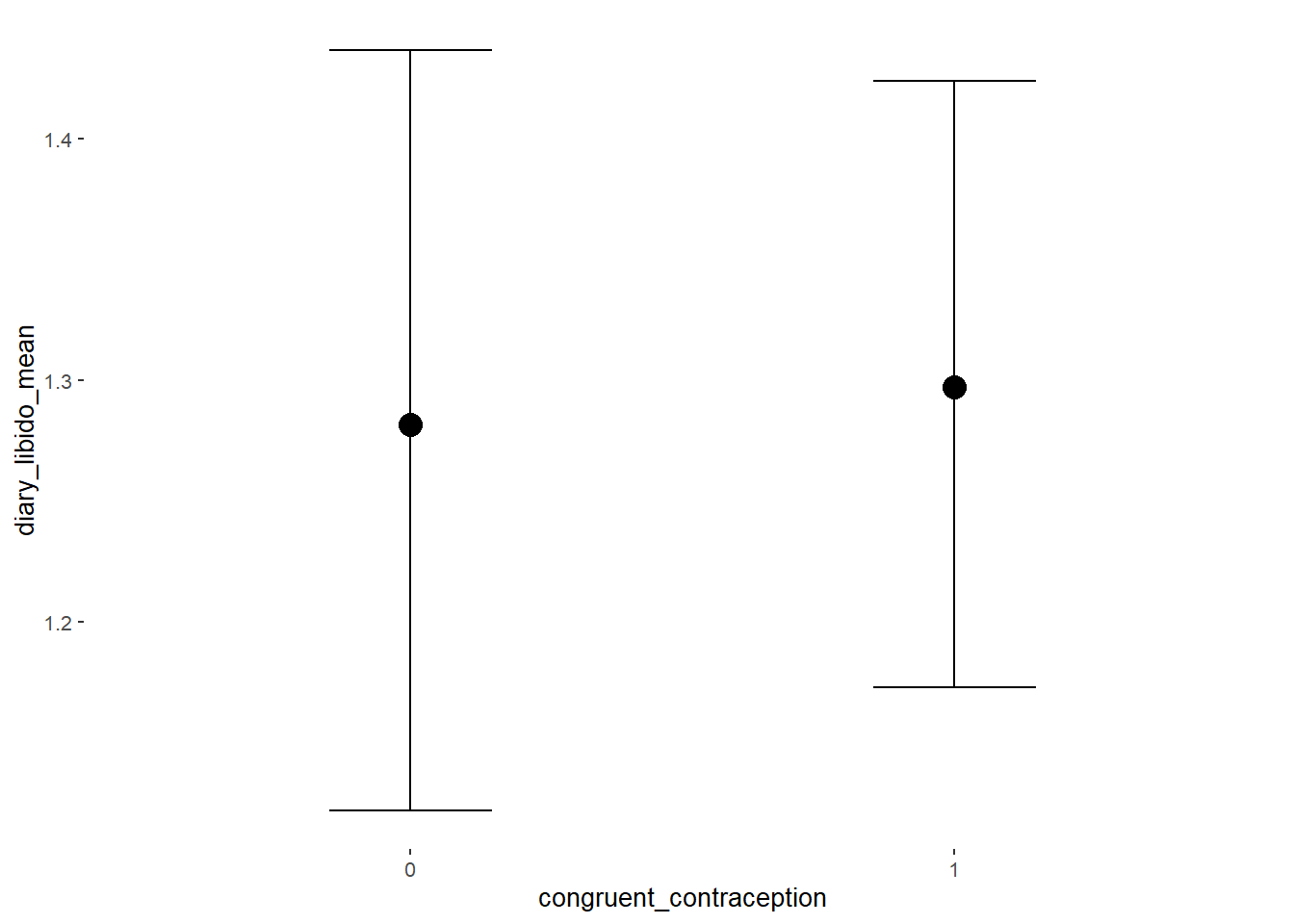
## Formula: diary_libido_mean ~ contraception_hormonal * congruent_contraception
## Data: data (Number of observations: 632)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept 1.24 0.05 1.16 1.32 1.00 2486
## contraception_hormonalyes -0.00 0.07 -0.12 0.12 1.00 2076
## congruent_contraception1 0.08 0.06 -0.02 0.18 1.00 2440
## contraception_hormonalyes:congruent_contraception1 -0.03 0.09 -0.18 0.12 1.00 2006
## Tail_ESS
## Intercept 2557
## contraception_hormonalyes 2537
## congruent_contraception1 2560
## contraception_hormonalyes:congruent_contraception1 2333
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.55 0.02 0.52 0.57 1.00 2952 2594
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_congruency_libido, range = c(-0.06, 0.06), ci = 0.90,
parameters = "contraception"))## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.81). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## Picking joint bandwidth of 0.0104## Warning: Removed 1197 rows containing non-finite values (stat_density_ridges).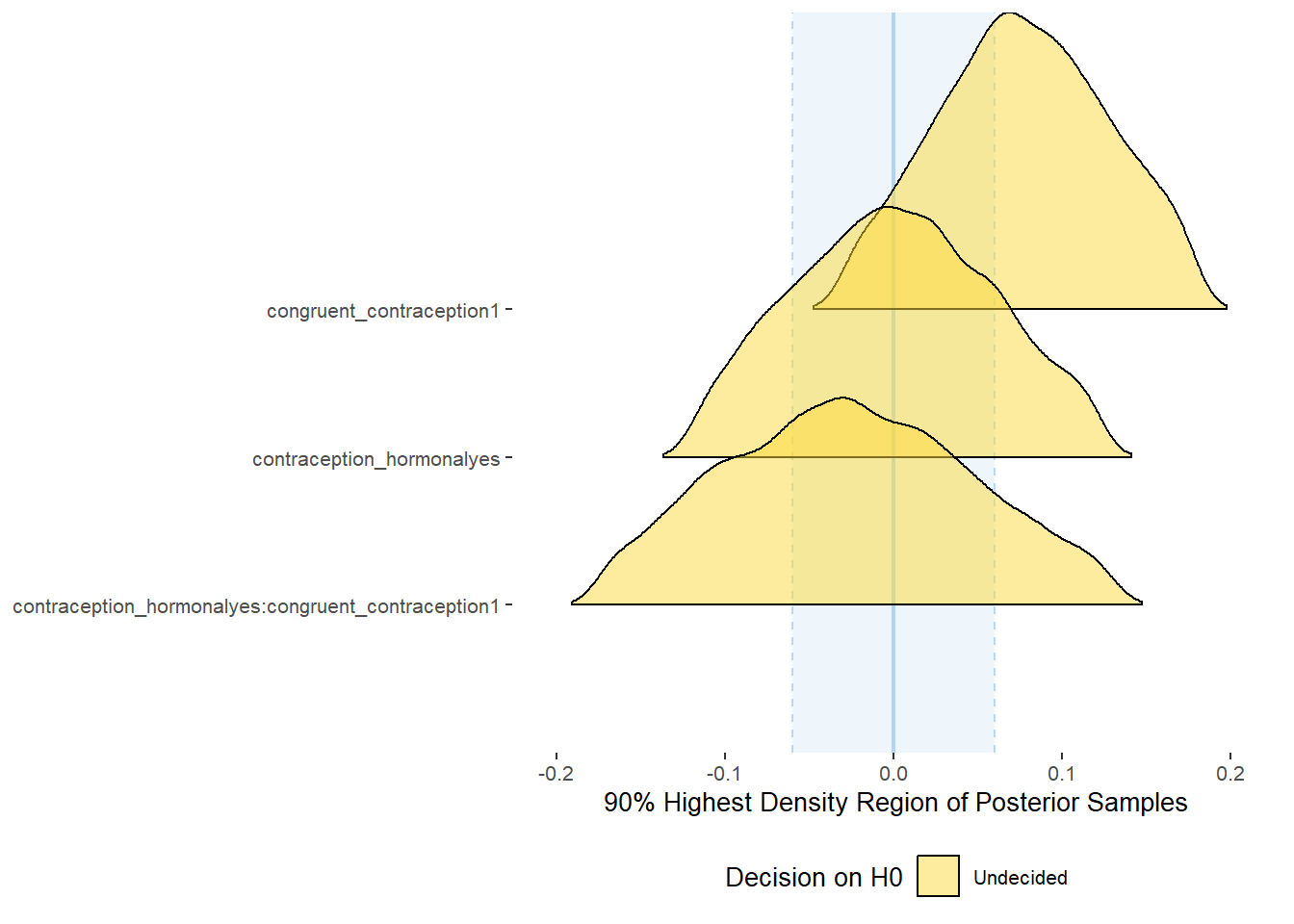
equivalence_test(m_congruency_libido, range = c(-0.06, 0.06), ci = 0.90,
parameters = "contraception")## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.81). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## # A tibble: 3 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.06 0.06 0.658 Undecided -0.119 0.122 fixed conditio~
## 2 b_congruent_co~ 0.9 -0.06 0.06 0.373 Undecided -0.0286 0.178 fixed conditio~
## 3 b_contraceptio~ 0.9 -0.06 0.06 0.534 Undecided -0.174 0.131 fixed conditio~Forest Plot for Effect Sizes
m_congruency_libido %>%
spread_draws(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`) %>%
pivot_longer(cols = c(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "ontraception",
"Contraception", NA),
condition = ifelse(condition == "b_contraception_hormonalyes",
"Hormonal Contraception",
ifelse(condition == "b_congruent_contraception1",
"Congruent Contraception",
ifelse(condition == "b_contraception_hormonalyes:congruent_contraception1",
"Interaction Hormonal Contracpetion and Congruent Contraception",
condition))),
condition = factor(condition, levels = rev(c("Hormonal Contraception",
"Congruent Contraception",
"Interaction Hormonal Contracpetion and Congruent Contraception")))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.06))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.06, 0.06), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors") +
xlim (-0.6, 0.6)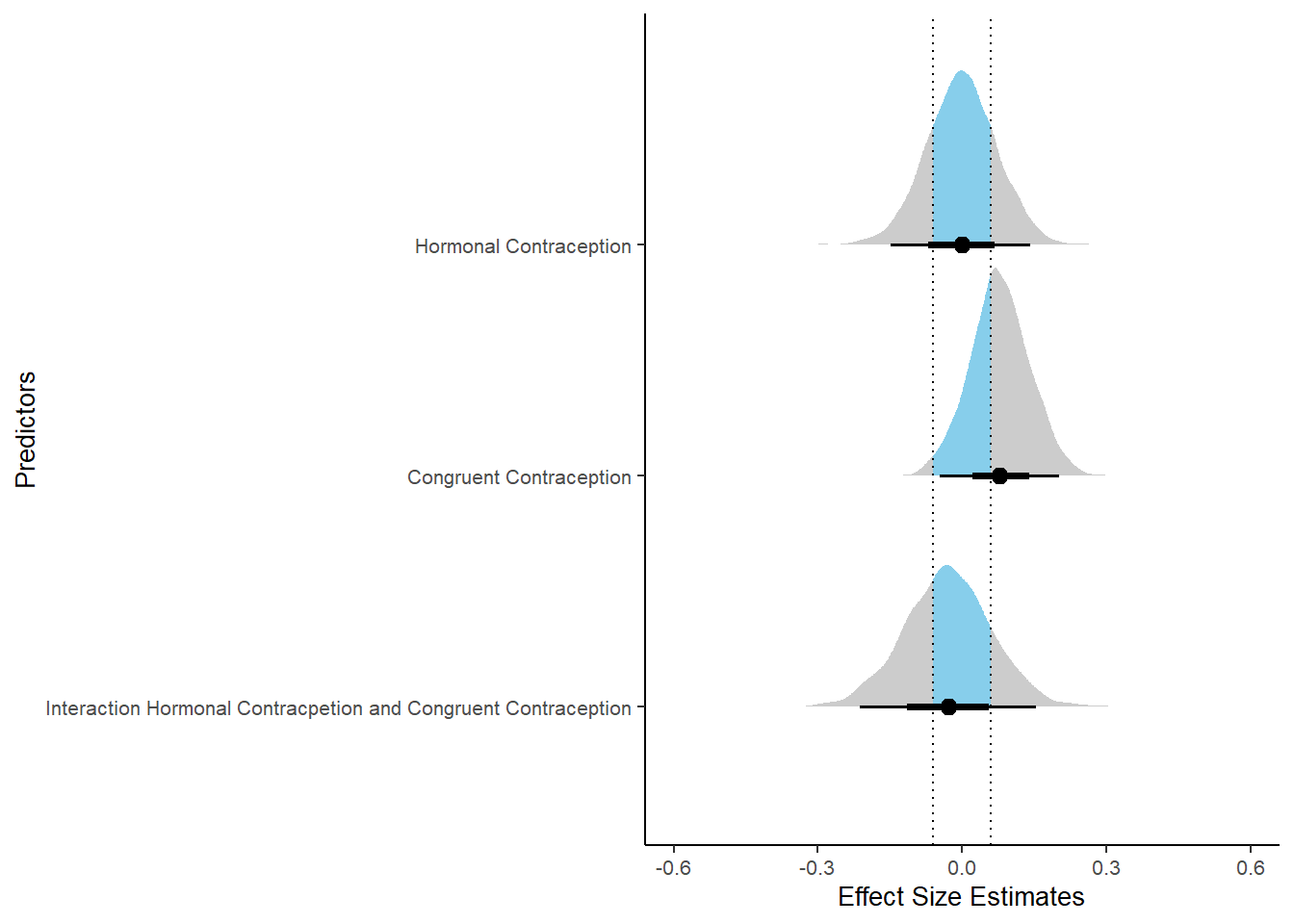
Sexual Frequency (Penetrative Intercourse)
Model
m_congruency_sexfreqpen = brm(diary_sex_active_sex_sum ~ offset(log(number_of_days)) +
contraception_hormonal * congruent_contraception,
data = data, family = poisson(),
file = "m_congruency_sexfreqpen")## Warning: Rows containing NAs were excluded from the model.## Compiling Stan program...## Start sampling##
## SAMPLING FOR MODEL 'c63cc6f90adde557ca52c07be9ae7367' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 0 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.933 seconds (Warm-up)
## Chain 1: 0.911 seconds (Sampling)
## Chain 1: 1.844 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL 'c63cc6f90adde557ca52c07be9ae7367' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 0 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 0.912 seconds (Warm-up)
## Chain 2: 0.94 seconds (Sampling)
## Chain 2: 1.852 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL 'c63cc6f90adde557ca52c07be9ae7367' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 0 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 0.99 seconds (Warm-up)
## Chain 3: 1.03 seconds (Sampling)
## Chain 3: 2.02 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL 'c63cc6f90adde557ca52c07be9ae7367' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 0 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 0.91 seconds (Warm-up)
## Chain 4: 0.96 seconds (Sampling)
## Chain 4: 1.87 seconds (Total)
## Chain 4:Summary
## Family: poisson
## Links: mu = log
## Formula: diary_sex_active_sex_sum ~ offset(log(number_of_days)) + contraception_hormonal * congruent_contraception
## Data: data (Number of observations: 622)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept -1.87 0.03 -1.93 -1.82 1.00 1883
## contraception_hormonalyes 0.20 0.05 0.12 0.27 1.00 1682
## congruent_contraception1 0.09 0.04 0.02 0.15 1.00 1774
## contraception_hormonalyes:congruent_contraception1 -0.09 0.06 -0.18 0.01 1.00 1535
## Tail_ESS
## Intercept 2465
## contraception_hormonalyes 2151
## congruent_contraception1 2486
## contraception_hormonalyes:congruent_contraception1 1891
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_congruency_sexfreqpen, range = c(-0.05, 0.05), ci = 0.90,
parameters = "contraception"))## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.81), b_contraception_hormonalyes:congruent_contraception1 and b_congruent_contraception1 (r = 0.71). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## Picking joint bandwidth of 0.00654## Warning: Removed 1197 rows containing non-finite values (stat_density_ridges).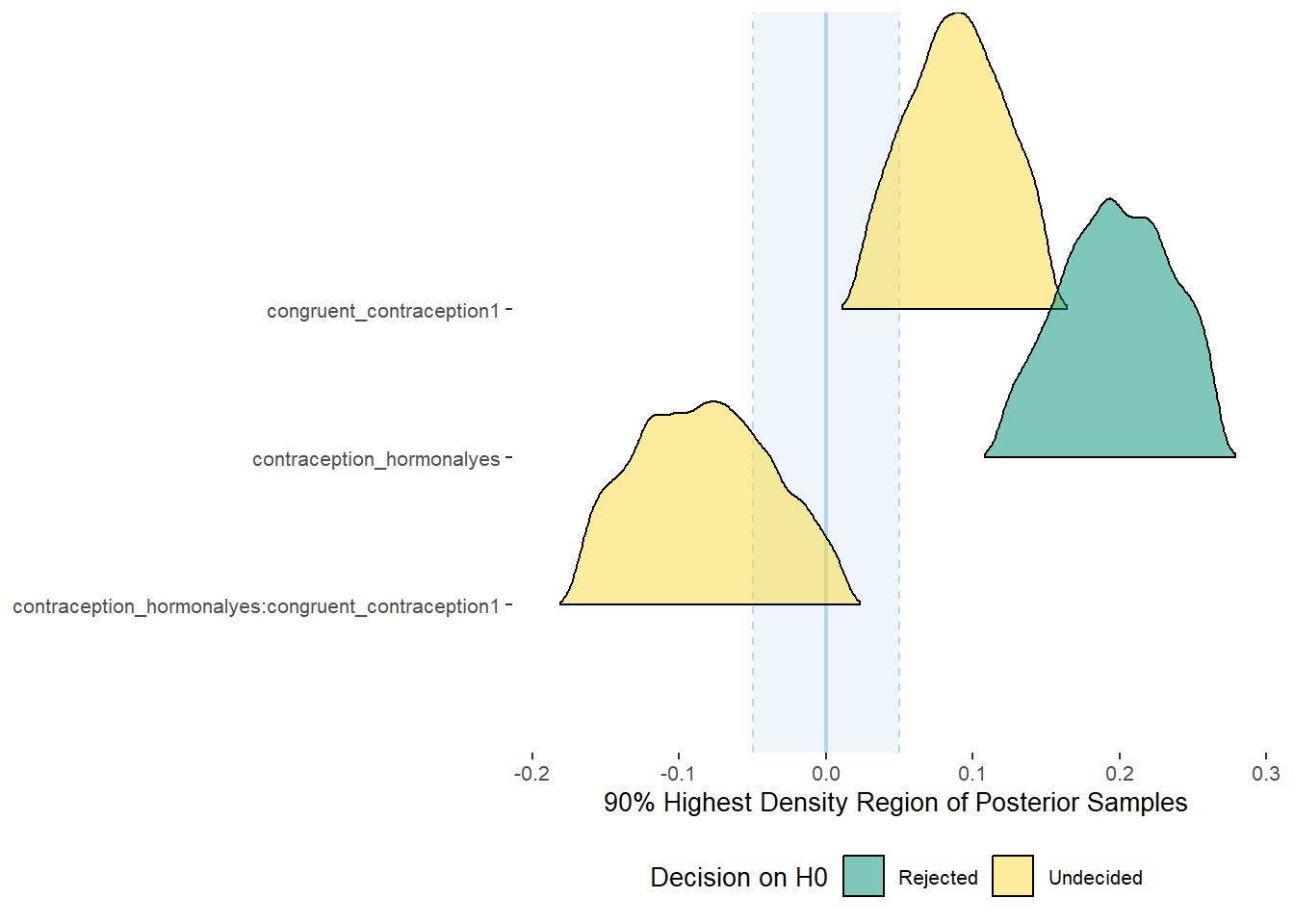
equivalence_test(m_congruency_sexfreqpen, range = c(-0.05, 0.05), ci = 0.90,
parameters = "contraception")## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.81), b_contraception_hormonalyes:congruent_contraception1 and b_congruent_contraception1 (r = 0.71). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## # A tibble: 3 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.05 0.05 0 Rejected 0.119 0.266 fixed conditio~
## 2 b_congruent_co~ 0.9 -0.05 0.05 0.134 Undecided 0.0225 0.152 fixed conditio~
## 3 b_contraceptio~ 0.9 -0.05 0.05 0.253 Undecided -0.170 0.0126 fixed conditio~Plots
conditional_effects(m_congruency_sexfreqpen,
effects = "contraception_hormonal:congruent_contraception",
conditions = data.frame(number_of_days = 1))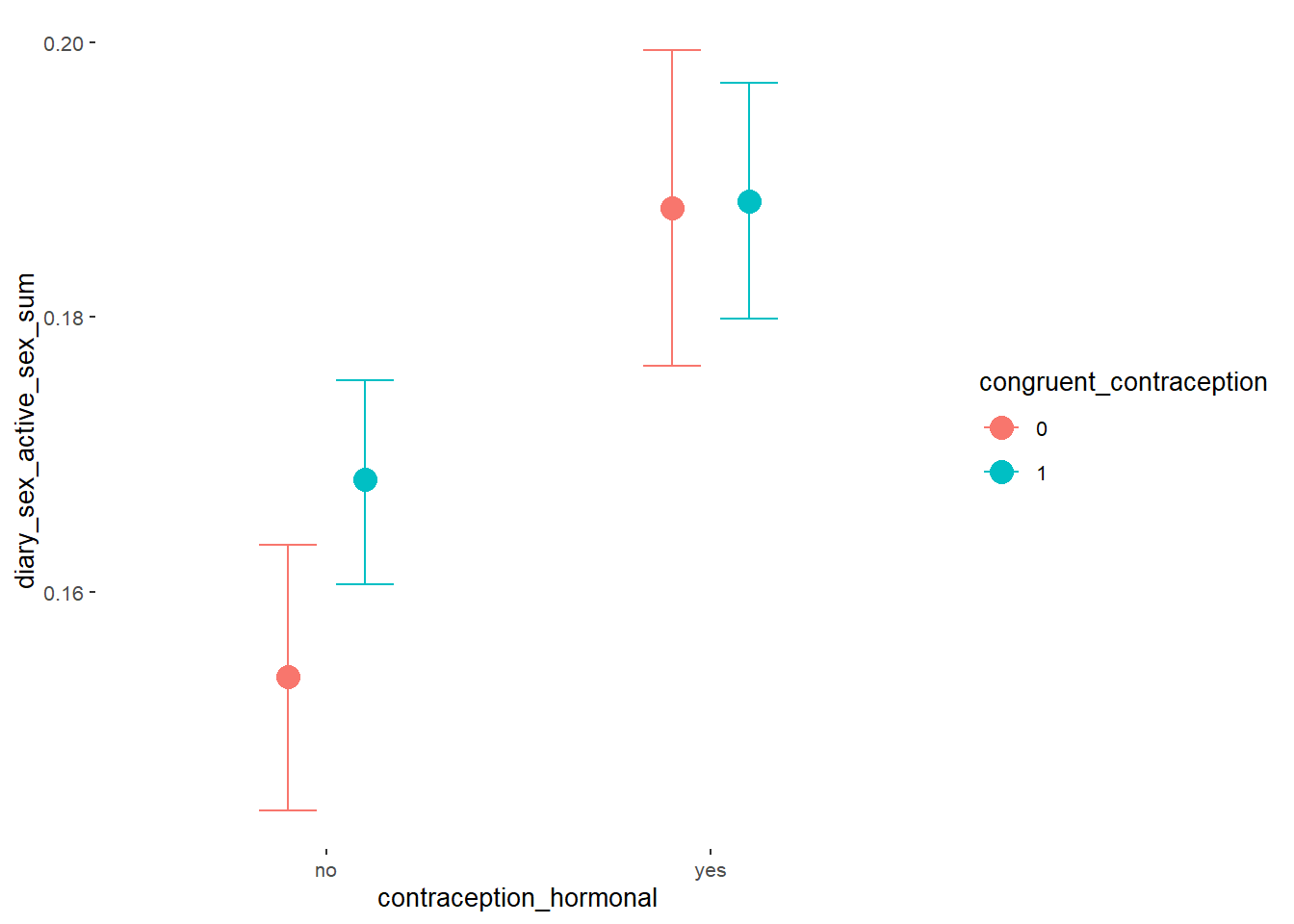
Forest Plot for Effect Sizes
m_congruency_sexfreqpen %>%
spread_draws(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`) %>%
pivot_longer(cols = c(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "ontraception",
"Contraception", NA),
condition = ifelse(condition == "b_contraception_hormonalyes",
"Hormonal Contraception",
ifelse(condition == "b_congruent_contraception1",
"Congruent Contraception",
ifelse(condition == "b_contraception_hormonalyes:congruent_contraception1",
"Interaction Hormonal Contracpetion and Congruent Contraception",
condition))),
condition = factor(condition, levels = rev(c("Hormonal Contraception",
"Congruent Contraception",
"Interaction Hormonal Contracpetion and Congruent Contraception")))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.05))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.05, 0.051), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")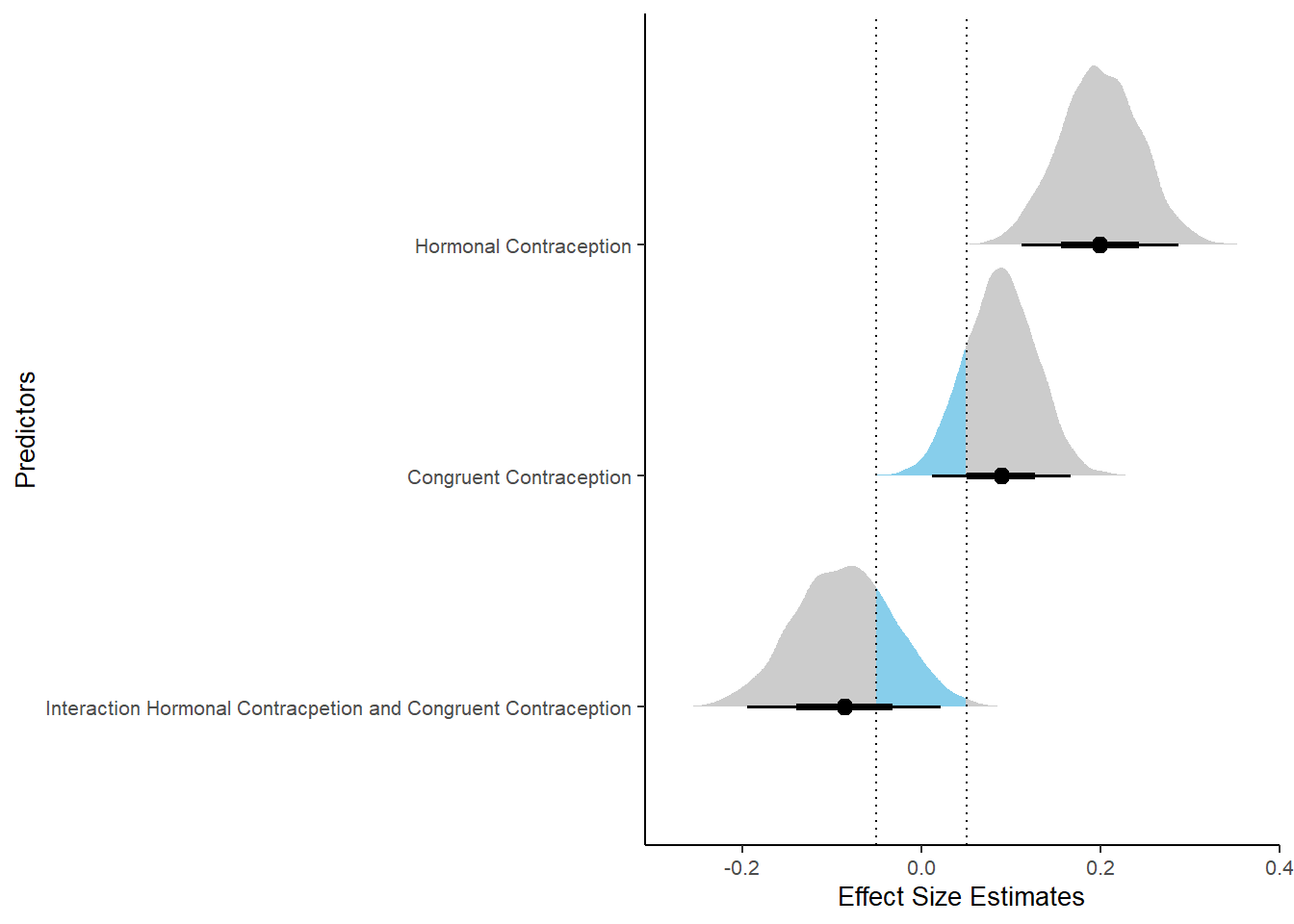
Masturbation Frequency
Model
Summary
## Family: poisson
## Links: mu = log
## Formula: diary_masturbation_sum ~ offset(log(number_of_days)) + contraception_hormonal * congruent_contraception
## Data: data (Number of observations: 622)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept -2.09 0.04 -2.15 -2.03 1.00 2289
## contraception_hormonalyes -0.37 0.06 -0.46 -0.27 1.00 1814
## congruent_contraception1 0.11 0.04 0.04 0.18 1.00 2096
## contraception_hormonalyes:congruent_contraception1 0.01 0.07 -0.10 0.13 1.00 1727
## Tail_ESS
## Intercept 2730
## contraception_hormonalyes 2328
## congruent_contraception1 2512
## contraception_hormonalyes:congruent_contraception1 2162
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_congruency_masfreq, range = c(-0.05, 0.05), ci = 0.90,
parameters = "contraception"))## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.82). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## Picking joint bandwidth of 0.00789## Warning: Removed 1197 rows containing non-finite values (stat_density_ridges).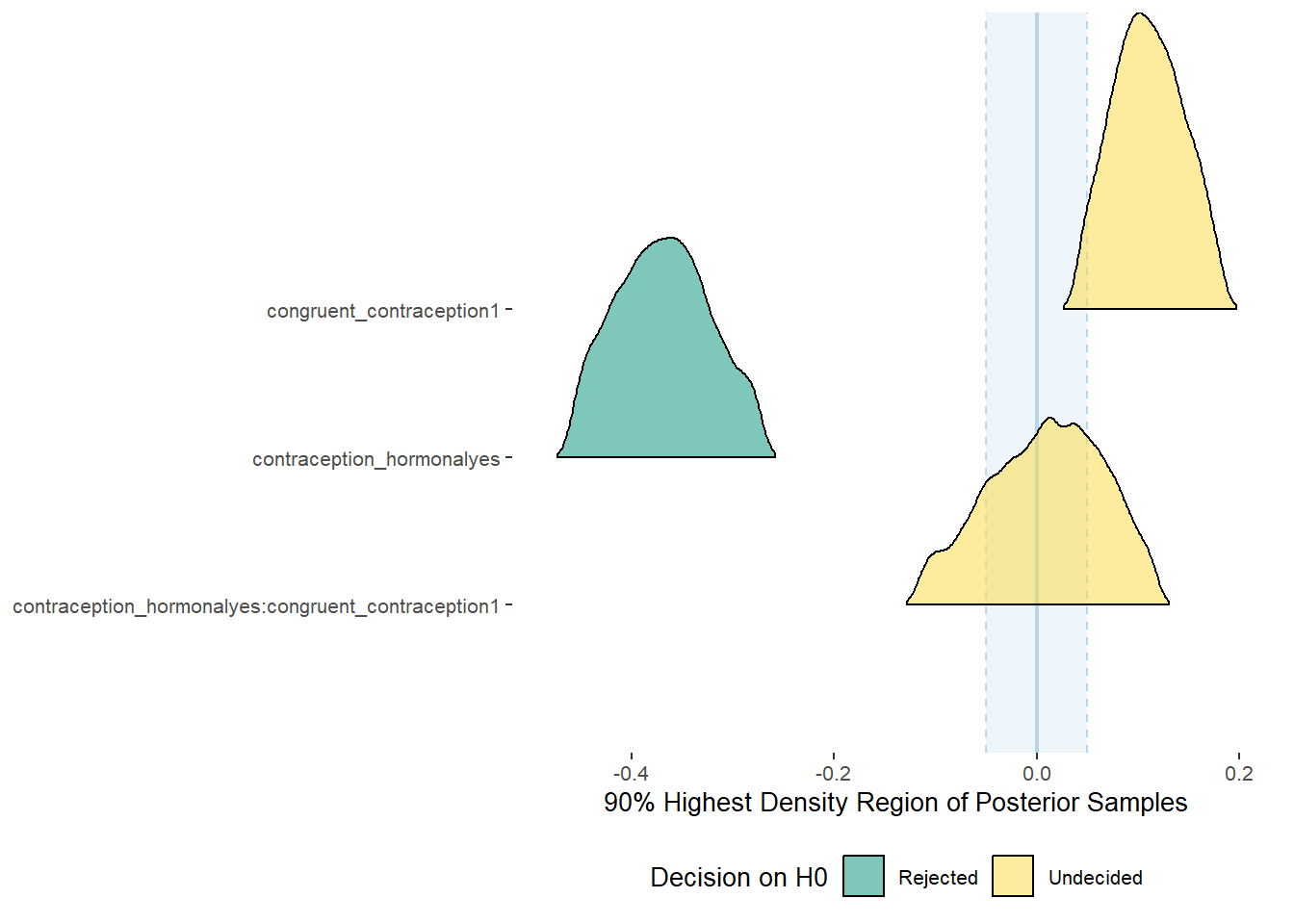
equivalence_test(m_congruency_masfreq, range = c(-0.05, 0.05), ci = 0.90,
parameters = "contraception")## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.82). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## # A tibble: 3 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.05 0.05 0 Rejected -0.460 -0.271 fixed conditio~
## 2 b_congruent_co~ 0.9 -0.05 0.05 0.0339 Undecided 0.0416 0.183 fixed conditio~
## 3 b_contraceptio~ 0.9 -0.05 0.05 0.571 Undecided -0.117 0.118 fixed conditio~Plots
conditional_effects(m_congruency_masfreq,
effects = "contraception_hormonal:congruent_contraception",
conditions = data.frame(number_of_days = 1))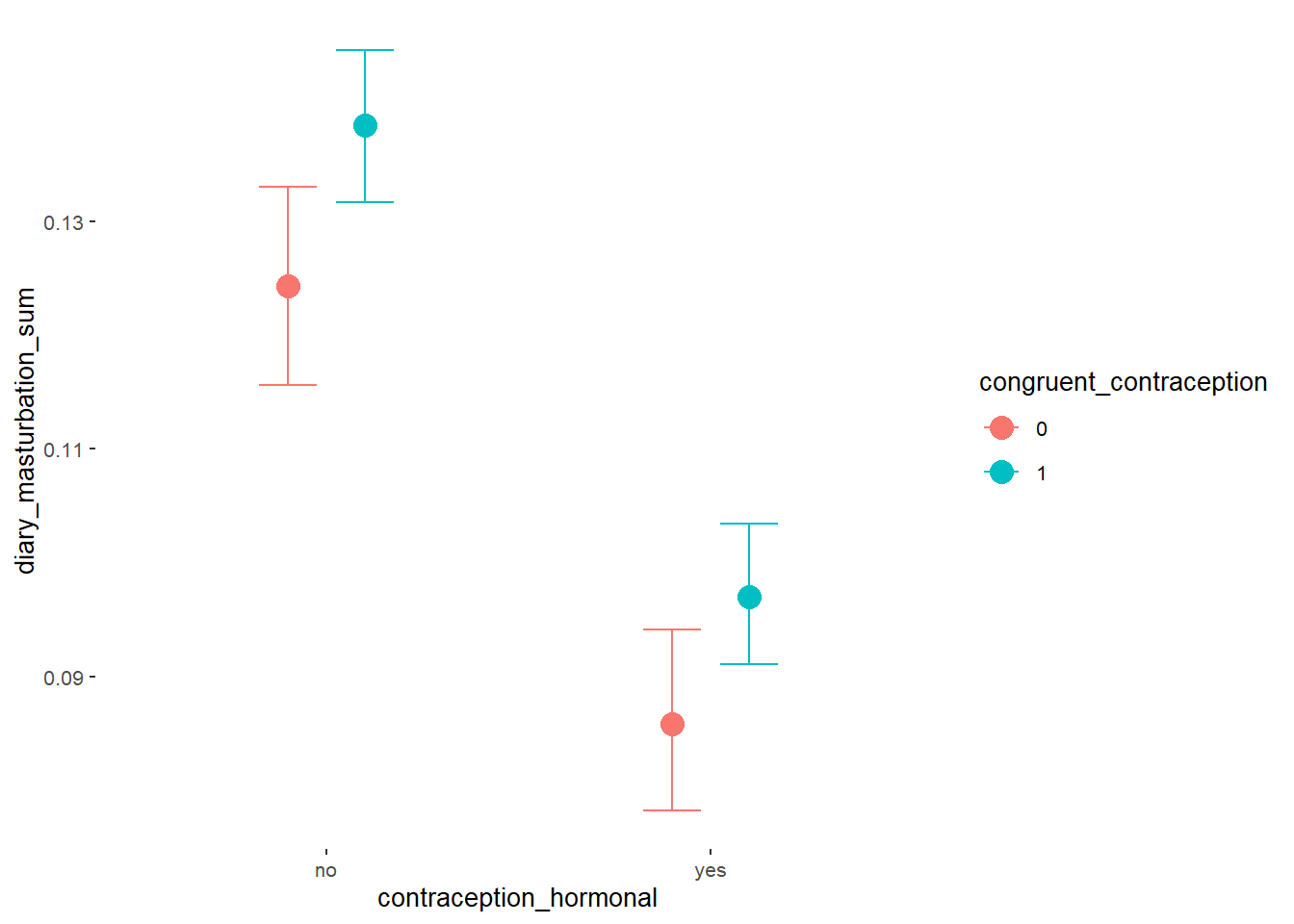
Forest Plot for Effect Sizes
m_congruency_masfreq %>%
spread_draws(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`) %>%
pivot_longer(cols = c(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "ontraception",
"Contraception", NA),
condition = ifelse(condition == "b_contraception_hormonalyes",
"Hormonal Contraception",
ifelse(condition == "b_congruent_contraception1",
"Congruent Contraception",
ifelse(condition == "b_contraception_hormonalyes:congruent_contraception1",
"Interaction Hormonal Contracpetion and Congruent Contraception",
condition))),
condition = factor(condition, levels = rev(c("Hormonal Contraception",
"Congruent Contraception",
"Interaction Hormonal Contracpetion and Congruent Contraception")))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.05))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.05, 0.05), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")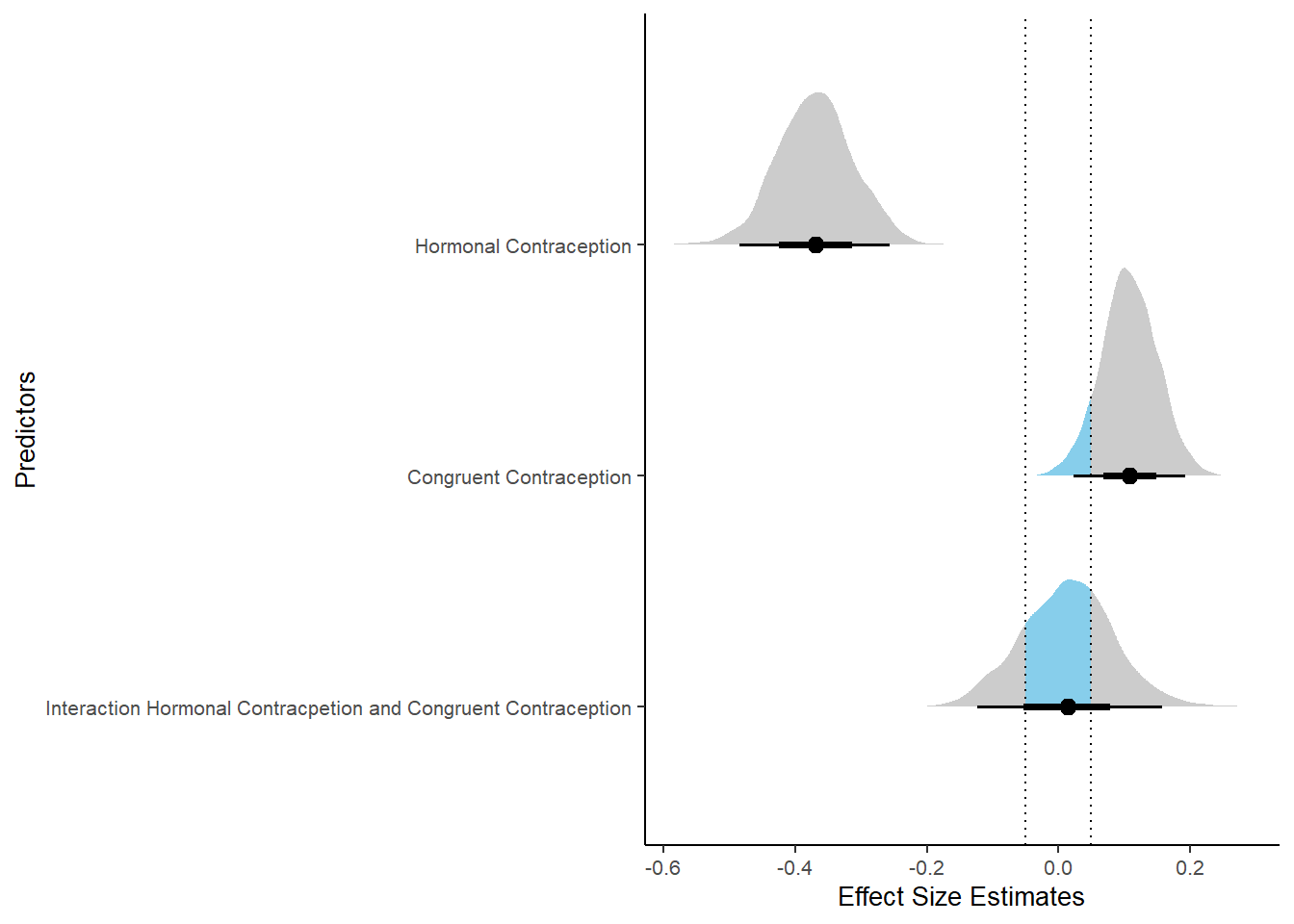
Controlled Models: Effects of (In)Congruenct HC Use
Attractiveness of Partner
Model
m_congruency_atrr_controlled = brm(attractiveness_partner ~
contraception_hormonal * congruent_contraception +
age + net_income + relationship_duration_factor +
education_years +
bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open +
religiosity,
data = data, family = gaussian(),
file = "m_congruency_atrr_controlled")Summary
## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: attractiveness_partner ~ contraception_hormonal * congruent_contraception + age + net_income + relationship_duration_factor + education_years + bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open + religiosity
## Data: data (Number of observations: 774)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept 3.21 0.37 2.59 3.80 1.00 4841
## contraception_hormonalyes 0.14 0.09 -0.01 0.29 1.00 3434
## congruent_contraception1 0.10 0.08 -0.03 0.23 1.00 3261
## age -0.00 0.01 -0.01 0.01 1.00 4709
## net_incomeeuro_500_1000 0.05 0.07 -0.07 0.16 1.00 3857
## net_incomeeuro_1000_2000 0.15 0.09 0.01 0.30 1.00 3699
## net_incomeeuro_2000_3000 0.19 0.12 -0.02 0.39 1.00 3928
## net_incomeeuro_gt_3000 0.18 0.21 -0.17 0.53 1.00 4157
## net_incomedont_tell -0.00 0.18 -0.30 0.29 1.00 4275
## relationship_duration_factorPartnered_upto28months 0.12 0.08 -0.01 0.24 1.00 3956
## relationship_duration_factorPartnered_upto52months -0.02 0.08 -0.15 0.11 1.00 3820
## relationship_duration_factorPartnered_morethan52months -0.12 0.08 -0.26 0.02 1.00 3703
## education_years 0.01 0.01 -0.00 0.02 1.00 5536
## bfi_extra 0.05 0.04 -0.01 0.11 1.00 4881
## bfi_neuro 0.00 0.04 -0.06 0.07 1.00 4395
## bfi_agree 0.10 0.05 0.03 0.18 1.00 4441
## bfi_consc 0.03 0.04 -0.04 0.10 1.00 5647
## bfi_open 0.06 0.05 -0.01 0.14 1.00 5430
## religiosity -0.00 0.02 -0.03 0.03 1.00 4886
## contraception_hormonalyes:congruent_contraception1 -0.09 0.11 -0.28 0.09 1.00 3107
## Tail_ESS
## Intercept 3175
## contraception_hormonalyes 2849
## congruent_contraception1 3133
## age 3701
## net_incomeeuro_500_1000 3299
## net_incomeeuro_1000_2000 2937
## net_incomeeuro_2000_3000 3154
## net_incomeeuro_gt_3000 3170
## net_incomedont_tell 2884
## relationship_duration_factorPartnered_upto28months 3040
## relationship_duration_factorPartnered_upto52months 3060
## relationship_duration_factorPartnered_morethan52months 3061
## education_years 3286
## bfi_extra 2971
## bfi_neuro 3347
## bfi_agree 3322
## bfi_consc 3084
## bfi_open 2807
## religiosity 3121
## contraception_hormonalyes:congruent_contraception1 2994
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.73 0.02 0.70 0.76 1.00 5358 2588
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_congruency_atrr_controlled, range = c(-0.07, 0.07), ci = 0.90,
parameters = "contraception"))## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.79), b_contraception_hormonalyes:congruent_contraception1 and b_congruent_contraception1 (r = 0.7). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## Picking joint bandwidth of 0.0132## Warning: Removed 1197 rows containing non-finite values (stat_density_ridges).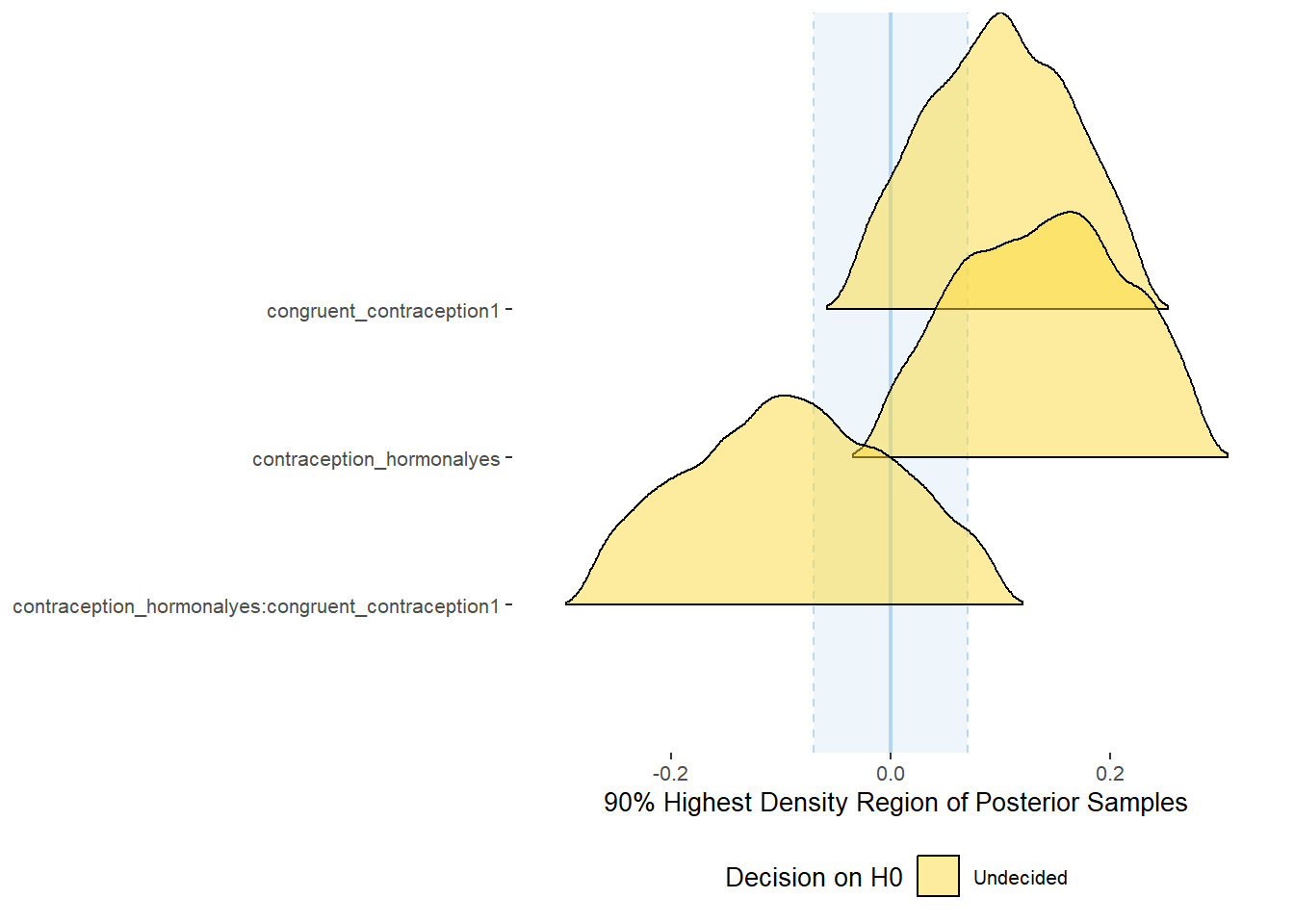
equivalence_test(m_congruency_atrr_controlled, range = c(-0.07, 0.07), ci = 0.90,
parameters = "contraception")## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.79), b_contraception_hormonalyes:congruent_contraception1 and b_congruent_contraception1 (r = 0.7). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## # A tibble: 3 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.07 0.07 0.200 Undecided -0.0107 0.284 fixed conditio~
## 2 b_congruent_co~ 0.9 -0.07 0.07 0.340 Undecided -0.0350 0.228 fixed conditio~
## 3 b_contraceptio~ 0.9 -0.07 0.07 0.383 Undecided -0.274 0.0992 fixed conditio~Forest Plot for Effect Sizes
m_congruency_atrr_controlled %>%
spread_draws(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity) %>%
pivot_longer(cols = c(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_relationship_duration_factor",
"Relationship Duration",
ifelse(condition %contains% "b_net_income",
"Income",
NA)),
group = ifelse(condition %contains% "ontraception",
"Contraception", group),
condition = ifelse(condition == "b_contraception_hormonalyes",
"Hormonal Contraception",
ifelse(condition == "b_congruent_contraception1",
"Congruent Contraception",
ifelse(condition == "b_contraception_hormonalyes:congruent_contraception1",
"Interaction Hormonal Contracpetion and Congruent Contraception",
condition))),
condition = ifelse(condition == "b_age", "Age",
ifelse(condition == "b_net_incomeeuro_500_1000", "500-1000 Euro",
ifelse(condition == "b_net_incomeeuro_1000_2000", "1000-2000 Euro",
ifelse(condition == "b_net_incomeeuro_2000_3000", "2000-3000 Euro",
ifelse(condition == "b_net_incomeeuro_gt_3000", ">3000 Euro",
ifelse(condition == "b_net_incomedont_tell", "do not tell",
ifelse(condition == "b_relationship_duration_factorPartnered_upto28months",
"13-28 months",
ifelse(condition == "b_relationship_duration_factorPartnered_upto52months",
"29-52 months",
ifelse(condition == "b_relationship_duration_factorPartnered_morethan52months",
">52 months",
ifelse(condition == "b_education_years", "Years of Education",
ifelse(condition == "b_bfi_extra", "Extraversion",
ifelse(condition == "b_bfi_neuro", "Neuroticism",
ifelse(condition == "b_bfi_agree", "Agreeableness",
ifelse(condition == "b_bfi_consc", "Conscientiousness",
ifelse(condition == "b_bfi_open", "Openness",
ifelse(condition == "b_religiosity", "Religiosity",
condition)))))))))))))))),
group = ifelse(is.na(group), condition, group),
condition = factor(condition, levels = rev(c("Hormonal Contraception",
"Congruent Contraception",
"Interaction Hormonal Contracpetion and Congruent Contraception",
"Age",
"500-1000 Euro", "1000-2000 Euro",
"2000-3000 Euro", ">3000 Euro", "do not tell",
"13-28 months", "29-52 months",
">52 months",
"Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))),
group = factor(group, levels = c("Contraception", "Age", "Income",
"Relationship Duration","Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.07))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.07, 0.07), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")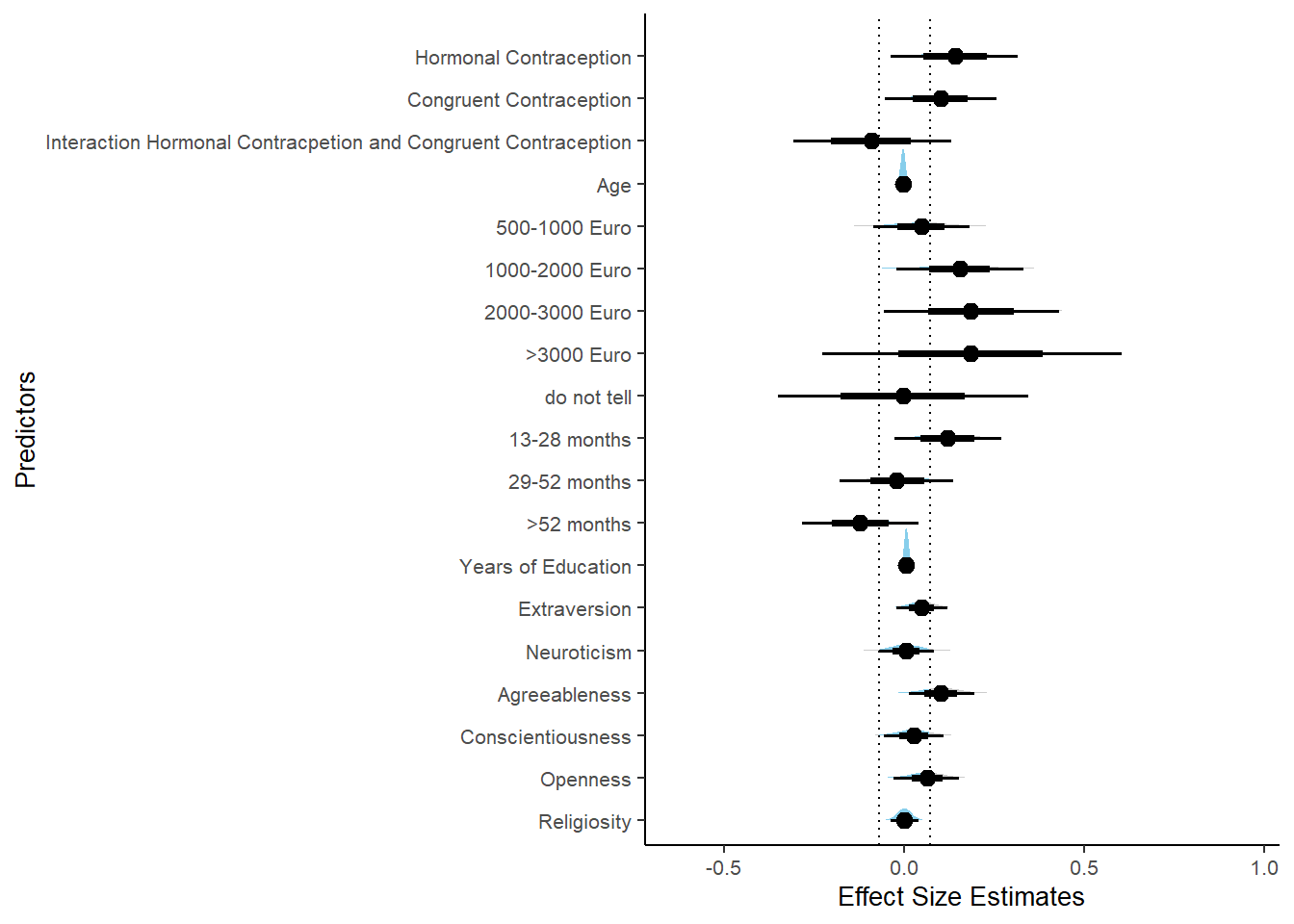
Relationship Satisfaction
Model
m_congruency_relsat_controlled = brm(relationship_satisfaction ~
contraception_hormonal * congruent_contraception +
age + net_income + relationship_duration_factor +
education_years +
bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open +
religiosity,
data = data, family = gaussian(),
file = "m_congruency_relsat_controlled")Summary
## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: relationship_satisfaction ~ contraception_hormonal * congruent_contraception + age + net_income + relationship_duration_factor + education_years + bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open + religiosity
## Data: data (Number of observations: 774)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept 3.31 0.22 2.96 3.68 1.00 5170
## contraception_hormonalyes 0.04 0.05 -0.05 0.12 1.00 3344
## congruent_contraception1 -0.07 0.04 -0.14 0.01 1.00 3427
## age -0.00 0.00 -0.01 0.00 1.00 4658
## net_incomeeuro_500_1000 0.07 0.04 0.00 0.13 1.00 3823
## net_incomeeuro_1000_2000 -0.01 0.05 -0.09 0.07 1.00 3900
## net_incomeeuro_2000_3000 0.03 0.07 -0.09 0.14 1.00 3449
## net_incomeeuro_gt_3000 0.11 0.12 -0.08 0.30 1.00 4589
## net_incomedont_tell -0.10 0.10 -0.27 0.07 1.00 4754
## relationship_duration_factorPartnered_upto28months 0.20 0.04 0.13 0.27 1.00 3757
## relationship_duration_factorPartnered_upto52months 0.15 0.05 0.08 0.22 1.00 3520
## relationship_duration_factorPartnered_morethan52months 0.13 0.05 0.05 0.20 1.00 3825
## education_years -0.00 0.00 -0.01 0.00 1.00 5870
## bfi_extra 0.02 0.02 -0.01 0.05 1.00 4186
## bfi_neuro 0.03 0.02 -0.01 0.07 1.00 4381
## bfi_agree -0.02 0.03 -0.07 0.02 1.00 4860
## bfi_consc 0.01 0.02 -0.04 0.05 1.00 5180
## bfi_open -0.01 0.03 -0.06 0.03 1.00 5037
## religiosity 0.03 0.01 0.01 0.05 1.00 4740
## contraception_hormonalyes:congruent_contraception1 0.03 0.06 -0.07 0.14 1.00 2955
## Tail_ESS
## Intercept 3333
## contraception_hormonalyes 2833
## congruent_contraception1 2906
## age 3556
## net_incomeeuro_500_1000 3101
## net_incomeeuro_1000_2000 3363
## net_incomeeuro_2000_3000 3233
## net_incomeeuro_gt_3000 3302
## net_incomedont_tell 3396
## relationship_duration_factorPartnered_upto28months 2670
## relationship_duration_factorPartnered_upto52months 3015
## relationship_duration_factorPartnered_morethan52months 3290
## education_years 3045
## bfi_extra 2997
## bfi_neuro 3294
## bfi_agree 3249
## bfi_consc 3217
## bfi_open 3099
## religiosity 2961
## contraception_hormonalyes:congruent_contraception1 2483
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.41 0.01 0.40 0.43 1.00 4750 3091
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_congruency_relsat_controlled, range = c(-0.04, 0.04), ci = 0.90,
parameters = "contraception"))## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.77). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## Picking joint bandwidth of 0.0073## Warning: Removed 1197 rows containing non-finite values (stat_density_ridges).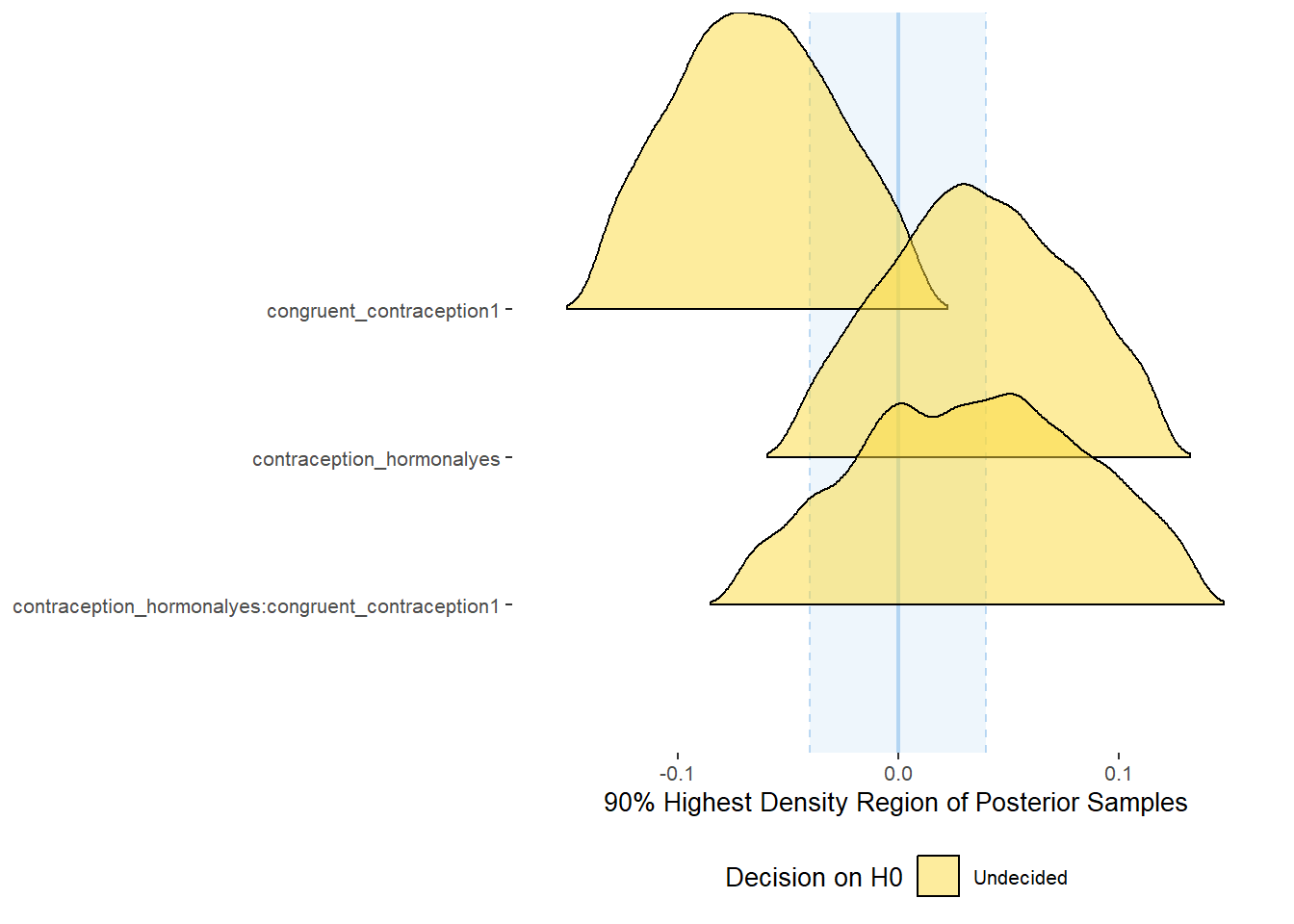
equivalence_test(m_congruency_relsat_controlled, range = c(-0.04, 0.04), ci = 0.90,
parameters = "contraception")## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.77). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## # A tibble: 3 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.04 0.04 0.520 Undecided -0.0470 0.119 fixed conditio~
## 2 b_congruent_co~ 0.9 -0.04 0.04 0.260 Undecided -0.137 0.00916 fixed conditio~
## 3 b_contraceptio~ 0.9 -0.04 0.04 0.459 Undecided -0.0731 0.136 fixed conditio~Forest Plot for Effect Sizes
m_congruency_relsat_controlled %>%
spread_draws(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity) %>%
pivot_longer(cols = c(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_relationship_duration_factor",
"Relationship Duration",
ifelse(condition %contains% "b_net_income",
"Income",
NA)),
group = ifelse(condition %contains% "ontraception",
"Contraception", group),
condition = ifelse(condition == "b_contraception_hormonalyes",
"Hormonal Contraception",
ifelse(condition == "b_congruent_contraception1",
"Congruent Contraception",
ifelse(condition == "b_contraception_hormonalyes:congruent_contraception1",
"Interaction Hormonal Contracpetion and Congruent Contraception",
condition))),
condition = ifelse(condition == "b_age", "Age",
ifelse(condition == "b_net_incomeeuro_500_1000", "500-1000 Euro",
ifelse(condition == "b_net_incomeeuro_1000_2000", "1000-2000 Euro",
ifelse(condition == "b_net_incomeeuro_2000_3000", "2000-3000 Euro",
ifelse(condition == "b_net_incomeeuro_gt_3000", ">3000 Euro",
ifelse(condition == "b_net_incomedont_tell", "do not tell",
ifelse(condition == "b_relationship_duration_factorPartnered_upto28months",
"13-28 months",
ifelse(condition == "b_relationship_duration_factorPartnered_upto52months",
"29-52 months",
ifelse(condition == "b_relationship_duration_factorPartnered_morethan52months",
">52 months",
ifelse(condition == "b_education_years", "Years of Education",
ifelse(condition == "b_bfi_extra", "Extraversion",
ifelse(condition == "b_bfi_neuro", "Neuroticism",
ifelse(condition == "b_bfi_agree", "Agreeableness",
ifelse(condition == "b_bfi_consc", "Conscientiousness",
ifelse(condition == "b_bfi_open", "Openness",
ifelse(condition == "b_religiosity", "Religiosity",
condition)))))))))))))))),
group = ifelse(is.na(group), condition, group),
condition = factor(condition, levels = rev(c("Hormonal Contraception",
"Congruent Contraception",
"Interaction Hormonal Contracpetion and Congruent Contraception",
"Age",
"500-1000 Euro", "1000-2000 Euro",
"2000-3000 Euro", ">3000 Euro", "do not tell",
"13-28 months", "29-52 months",
">52 months",
"Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))),
group = factor(group, levels = c("Contraception", "Age", "Income",
"Relationship Duration","Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.04))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.04, 0.04), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")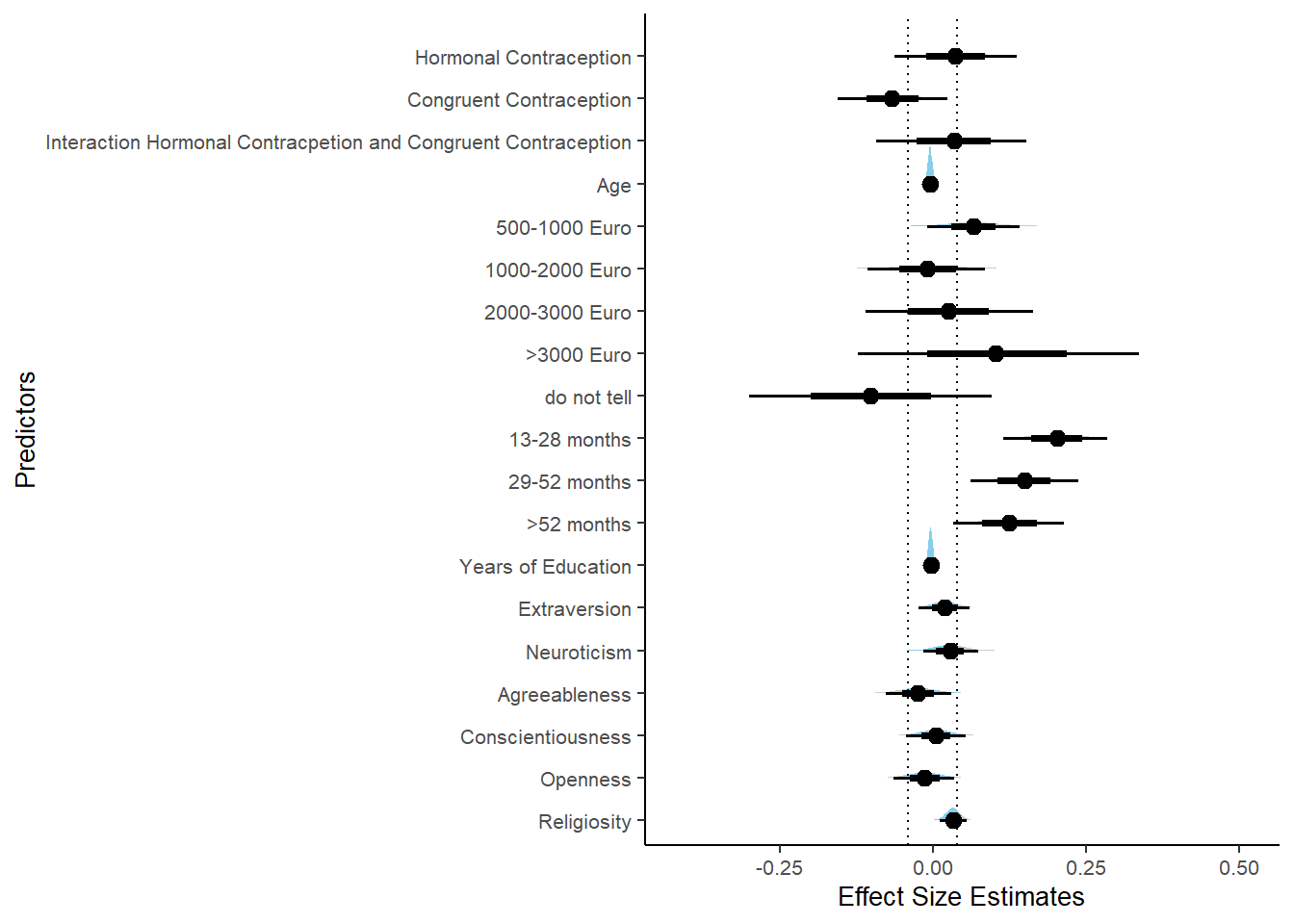
Sexual Satisfaction
Model
m_congruency_sexsat_controlled = brm(satisfaction_sexual_intercourse ~
contraception_hormonal * congruent_contraception +
age + net_income + relationship_duration_factor +
education_years +
bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open +
religiosity,
data = data, family = gaussian(),
file = "m_congruency_sexsat_controlled")Summary
## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: satisfaction_sexual_intercourse ~ contraception_hormonal * congruent_contraception + age + net_income + relationship_duration_factor + education_years + bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open + religiosity
## Data: data (Number of observations: 774)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept 3.18 0.55 2.28 4.07 1.00 4935
## contraception_hormonalyes 0.12 0.13 -0.10 0.33 1.00 3289
## congruent_contraception1 0.04 0.12 -0.15 0.23 1.00 3444
## age 0.01 0.01 -0.01 0.02 1.00 4130
## net_incomeeuro_500_1000 0.03 0.10 -0.13 0.18 1.00 3957
## net_incomeeuro_1000_2000 -0.08 0.13 -0.29 0.13 1.00 3414
## net_incomeeuro_2000_3000 -0.08 0.18 -0.37 0.21 1.00 4057
## net_incomeeuro_gt_3000 -0.26 0.30 -0.75 0.24 1.00 4579
## net_incomedont_tell -0.04 0.25 -0.46 0.36 1.00 4603
## relationship_duration_factorPartnered_upto28months -0.02 0.11 -0.19 0.16 1.00 4262
## relationship_duration_factorPartnered_upto52months -0.23 0.11 -0.42 -0.05 1.00 4104
## relationship_duration_factorPartnered_morethan52months -0.37 0.12 -0.56 -0.18 1.00 4017
## education_years -0.00 0.01 -0.02 0.01 1.00 5468
## bfi_extra 0.11 0.05 0.02 0.19 1.00 5191
## bfi_neuro -0.07 0.06 -0.17 0.02 1.00 4528
## bfi_agree 0.13 0.07 0.03 0.24 1.00 5064
## bfi_consc 0.14 0.06 0.04 0.23 1.00 5904
## bfi_open -0.10 0.06 -0.20 0.01 1.00 5260
## religiosity -0.01 0.03 -0.05 0.04 1.00 5701
## contraception_hormonalyes:congruent_contraception1 -0.01 0.17 -0.28 0.26 1.00 3163
## Tail_ESS
## Intercept 2946
## contraception_hormonalyes 2883
## congruent_contraception1 3135
## age 2864
## net_incomeeuro_500_1000 3187
## net_incomeeuro_1000_2000 2932
## net_incomeeuro_2000_3000 2994
## net_incomeeuro_gt_3000 3080
## net_incomedont_tell 2890
## relationship_duration_factorPartnered_upto28months 3210
## relationship_duration_factorPartnered_upto52months 2756
## relationship_duration_factorPartnered_morethan52months 2881
## education_years 2948
## bfi_extra 3176
## bfi_neuro 3091
## bfi_agree 2730
## bfi_consc 2967
## bfi_open 3048
## religiosity 3140
## contraception_hormonalyes:congruent_contraception1 3035
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 1.03 0.03 0.99 1.08 1.00 4869 3374
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_congruency_sexsat_controlled, range = c(-0.11, 0.11), ci = 0.90,
parameters = "contraception"))## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.78), b_contraception_hormonalyes:congruent_contraception1 and b_congruent_contraception1 (r = 0.72). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## Picking joint bandwidth of 0.0189## Warning: Removed 1197 rows containing non-finite values (stat_density_ridges).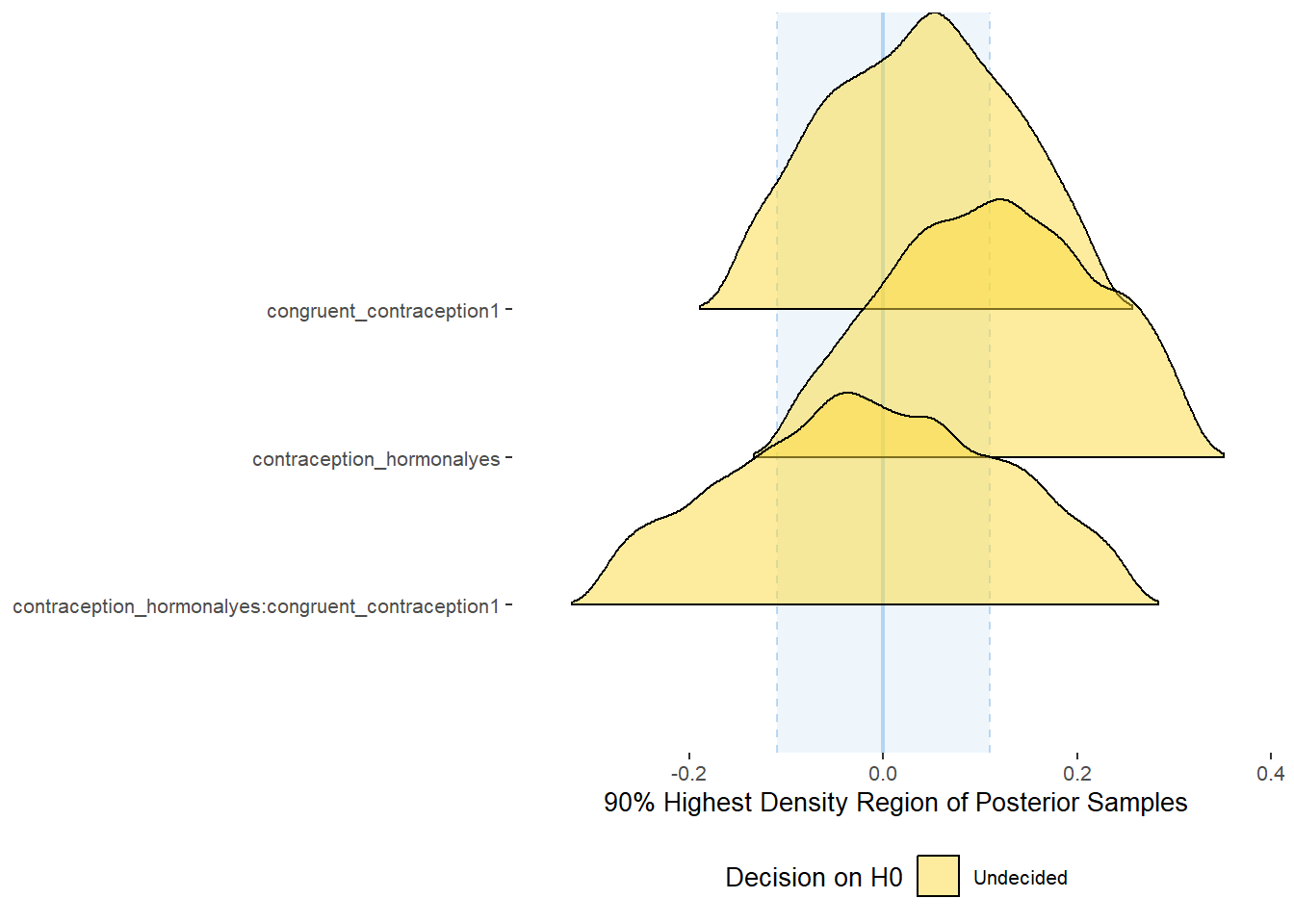
equivalence_test(m_congruency_sexsat_controlled, range = c(-0.11, 0.11), ci = 0.90,
parameters = "contraception")## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.78), b_contraception_hormonalyes:congruent_contraception1 and b_congruent_contraception1 (r = 0.72). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## # A tibble: 3 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.11 0.11 0.490 Undecided -0.102 0.317 fixed conditio~
## 2 b_congruent_co~ 0.9 -0.11 0.11 0.705 Undecided -0.155 0.222 fixed conditio~
## 3 b_contraceptio~ 0.9 -0.11 0.11 0.547 Undecided -0.290 0.253 fixed conditio~Forest Plot for Effect Sizes
m_congruency_sexsat_controlled %>%
spread_draws(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity) %>%
pivot_longer(cols = c(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_relationship_duration_factor",
"Relationship Duration",
ifelse(condition %contains% "b_net_income",
"Income",
NA)),
group = ifelse(condition %contains% "ontraception",
"Contraception", group),
condition = ifelse(condition == "b_contraception_hormonalyes",
"Hormonal Contraception",
ifelse(condition == "b_congruent_contraception1",
"Congruent Contraception",
ifelse(condition == "b_contraception_hormonalyes:congruent_contraception1",
"Interaction Hormonal Contracpetion and Congruent Contraception",
condition))),
condition = ifelse(condition == "b_age", "Age",
ifelse(condition == "b_net_incomeeuro_500_1000", "500-1000 Euro",
ifelse(condition == "b_net_incomeeuro_1000_2000", "1000-2000 Euro",
ifelse(condition == "b_net_incomeeuro_2000_3000", "2000-3000 Euro",
ifelse(condition == "b_net_incomeeuro_gt_3000", ">3000 Euro",
ifelse(condition == "b_net_incomedont_tell", "do not tell",
ifelse(condition == "b_relationship_duration_factorPartnered_upto28months",
"13-28 months",
ifelse(condition == "b_relationship_duration_factorPartnered_upto52months",
"29-52 months",
ifelse(condition == "b_relationship_duration_factorPartnered_morethan52months",
">52 months",
ifelse(condition == "b_education_years", "Years of Education",
ifelse(condition == "b_bfi_extra", "Extraversion",
ifelse(condition == "b_bfi_neuro", "Neuroticism",
ifelse(condition == "b_bfi_agree", "Agreeableness",
ifelse(condition == "b_bfi_consc", "Conscientiousness",
ifelse(condition == "b_bfi_open", "Openness",
ifelse(condition == "b_religiosity", "Religiosity",
condition)))))))))))))))),
group = ifelse(is.na(group), condition, group),
condition = factor(condition, levels = rev(c("Hormonal Contraception",
"Congruent Contraception",
"Interaction Hormonal Contracpetion and Congruent Contraception",
"Age",
"500-1000 Euro", "1000-2000 Euro",
"2000-3000 Euro", ">3000 Euro", "do not tell",
"13-28 months", "29-52 months",
">52 months",
"Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))),
group = factor(group, levels = c("Contraception", "Age", "Income",
"Relationship Duration","Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.11))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.11, 0.11), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")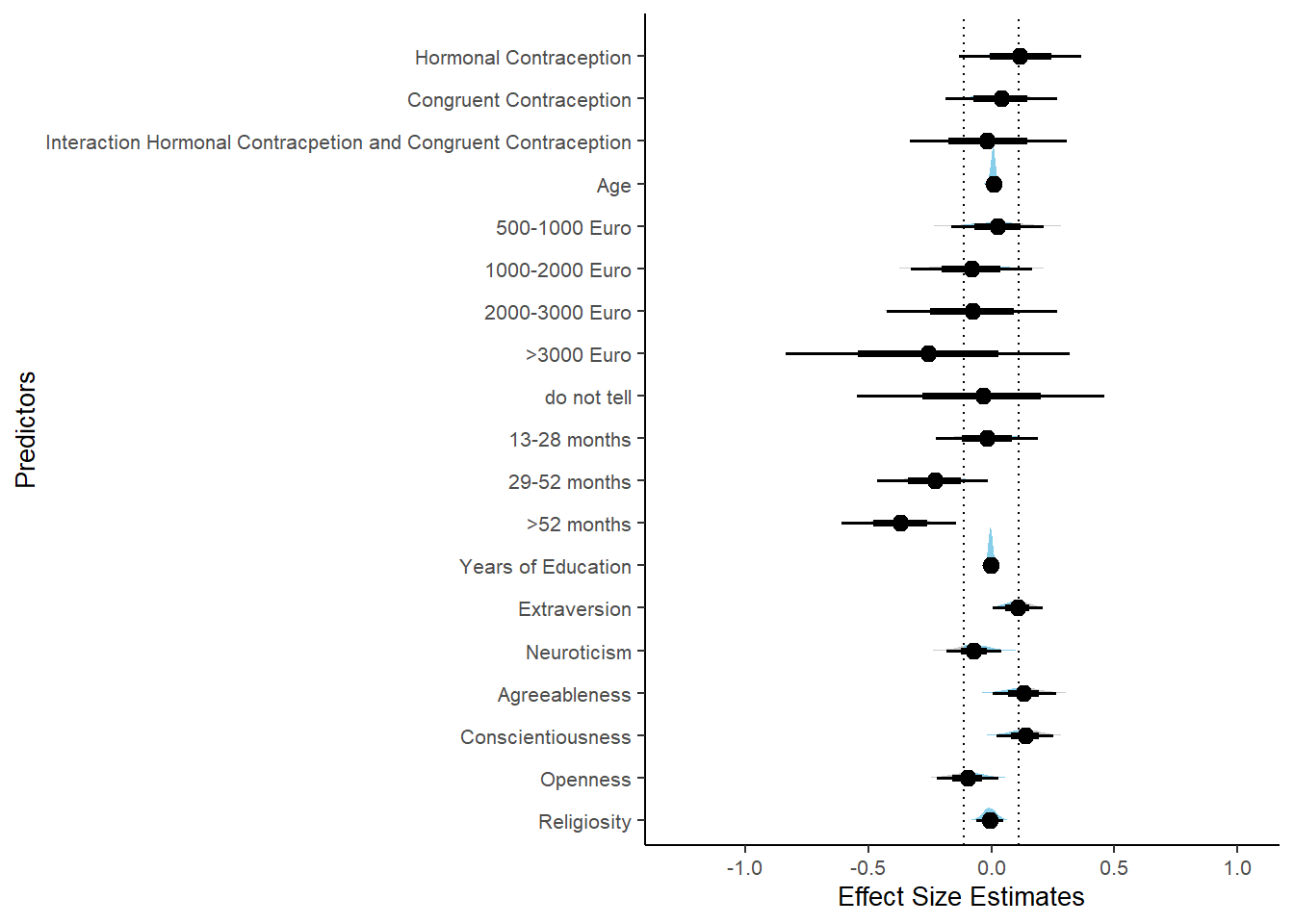
Libido
Model
m_congruency_libido_controlled = brm(diary_libido_mean ~
contraception_hormonal * congruent_contraception +
age + net_income + relationship_duration_factor +
education_years +
bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open +
religiosity,
data = data, family = gaussian(),
file = "m_congruency_libido_controlled")Summary
## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: diary_libido_mean ~ contraception_hormonal * congruent_contraception + age + net_income + relationship_duration_factor + education_years + bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open + religiosity
## Data: data (Number of observations: 632)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept 1.07 0.31 0.57 1.60 1.00 5382
## contraception_hormonalyes -0.02 0.07 -0.14 0.11 1.00 3571
## congruent_contraception1 0.02 0.06 -0.09 0.12 1.00 3601
## age -0.00 0.01 -0.01 0.00 1.00 4007
## net_incomeeuro_500_1000 0.12 0.05 0.03 0.21 1.00 3657
## net_incomeeuro_1000_2000 0.14 0.07 0.02 0.27 1.00 3164
## net_incomeeuro_2000_3000 0.14 0.10 -0.04 0.30 1.00 3487
## net_incomeeuro_gt_3000 0.02 0.18 -0.29 0.32 1.00 4150
## net_incomedont_tell 0.27 0.14 0.03 0.50 1.00 4255
## relationship_duration_factorPartnered_upto28months -0.11 0.06 -0.21 -0.02 1.00 3842
## relationship_duration_factorPartnered_upto52months -0.15 0.07 -0.26 -0.04 1.00 3830
## relationship_duration_factorPartnered_morethan52months -0.18 0.07 -0.29 -0.06 1.00 3838
## education_years -0.00 0.00 -0.01 0.01 1.00 5899
## bfi_extra 0.06 0.03 0.01 0.11 1.00 4656
## bfi_neuro -0.03 0.03 -0.08 0.03 1.00 4337
## bfi_agree 0.07 0.04 0.01 0.13 1.00 4542
## bfi_consc -0.09 0.03 -0.15 -0.04 1.00 5385
## bfi_open 0.09 0.04 0.03 0.15 1.00 5544
## religiosity -0.00 0.02 -0.03 0.02 1.00 5039
## contraception_hormonalyes:congruent_contraception1 0.04 0.09 -0.11 0.19 1.00 3167
## Tail_ESS
## Intercept 3690
## contraception_hormonalyes 2734
## congruent_contraception1 3274
## age 3216
## net_incomeeuro_500_1000 3272
## net_incomeeuro_1000_2000 2914
## net_incomeeuro_2000_3000 2962
## net_incomeeuro_gt_3000 2800
## net_incomedont_tell 2891
## relationship_duration_factorPartnered_upto28months 3163
## relationship_duration_factorPartnered_upto52months 3268
## relationship_duration_factorPartnered_morethan52months 3337
## education_years 3177
## bfi_extra 3376
## bfi_neuro 3014
## bfi_agree 3185
## bfi_consc 3310
## bfi_open 3153
## religiosity 3107
## contraception_hormonalyes:congruent_contraception1 2743
##
## Family Specific Parameters:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.53 0.02 0.51 0.56 1.00 4580 2911
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_congruency_libido_controlled, range = c(-0.06, 0.06), ci = 0.90,
parameters = "contraception"))## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.79). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## Picking joint bandwidth of 0.0105## Warning: Removed 1197 rows containing non-finite values (stat_density_ridges).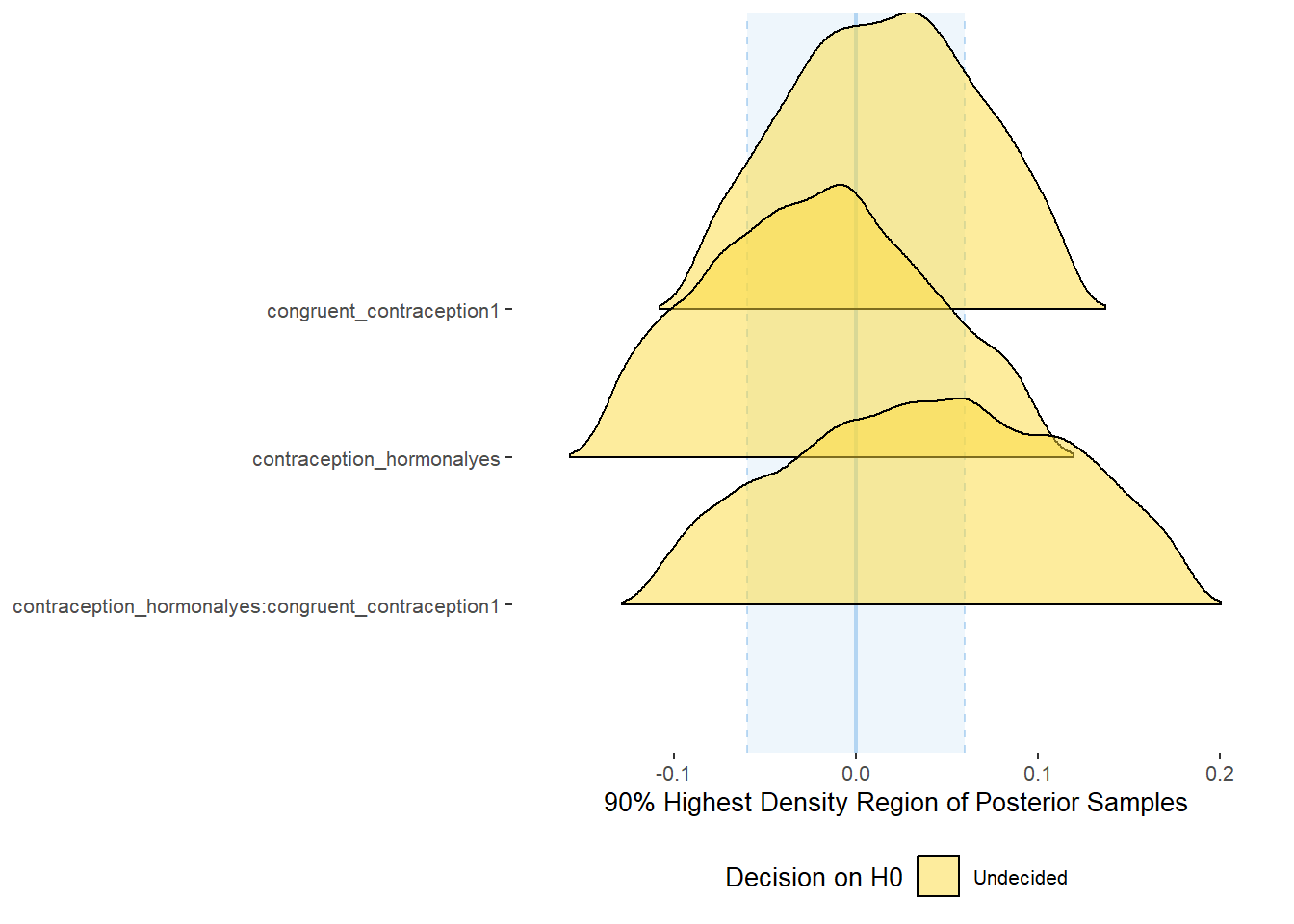
equivalence_test(m_congruency_libido_controlled, range = c(-0.06, 0.06), ci = 0.90,
parameters = "contraception")## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.79). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## # A tibble: 3 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.06 0.06 0.629 Undecided -0.138 0.102 fixed conditio~
## 2 b_congruent_co~ 0.9 -0.06 0.06 0.715 Undecided -0.0899 0.117 fixed conditio~
## 3 b_contraceptio~ 0.9 -0.06 0.06 0.490 Undecided -0.111 0.183 fixed conditio~Forest Plot for Effect Sizes
m_congruency_libido_controlled %>%
spread_draws(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity) %>%
pivot_longer(cols = c(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_relationship_duration_factor",
"Relationship Duration",
ifelse(condition %contains% "b_net_income",
"Income",
NA)),
group = ifelse(condition %contains% "ontraception",
"Contraception", group),
condition = ifelse(condition == "b_contraception_hormonalyes",
"Hormonal Contraception",
ifelse(condition == "b_congruent_contraception1",
"Congruent Contraception",
ifelse(condition == "b_contraception_hormonalyes:congruent_contraception1",
"Interaction Hormonal Contracpetion and Congruent Contraception",
condition))),
condition = ifelse(condition == "b_age", "Age",
ifelse(condition == "b_net_incomeeuro_500_1000", "500-1000 Euro",
ifelse(condition == "b_net_incomeeuro_1000_2000", "1000-2000 Euro",
ifelse(condition == "b_net_incomeeuro_2000_3000", "2000-3000 Euro",
ifelse(condition == "b_net_incomeeuro_gt_3000", ">3000 Euro",
ifelse(condition == "b_net_incomedont_tell", "do not tell",
ifelse(condition == "b_relationship_duration_factorPartnered_upto28months",
"13-28 months",
ifelse(condition == "b_relationship_duration_factorPartnered_upto52months",
"29-52 months",
ifelse(condition == "b_relationship_duration_factorPartnered_morethan52months",
">52 months",
ifelse(condition == "b_education_years", "Years of Education",
ifelse(condition == "b_bfi_extra", "Extraversion",
ifelse(condition == "b_bfi_neuro", "Neuroticism",
ifelse(condition == "b_bfi_agree", "Agreeableness",
ifelse(condition == "b_bfi_consc", "Conscientiousness",
ifelse(condition == "b_bfi_open", "Openness",
ifelse(condition == "b_religiosity", "Religiosity",
condition)))))))))))))))),
group = ifelse(is.na(group), condition, group),
condition = factor(condition, levels = rev(c("Hormonal Contraception",
"Congruent Contraception",
"Interaction Hormonal Contracpetion and Congruent Contraception",
"Age",
"500-1000 Euro", "1000-2000 Euro",
"2000-3000 Euro", ">3000 Euro", "do not tell",
"13-28 months", "29-52 months",
">52 months",
"Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))),
group = factor(group, levels = c("Contraception", "Age", "Income",
"Relationship Duration","Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.06))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.06, 0.06), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")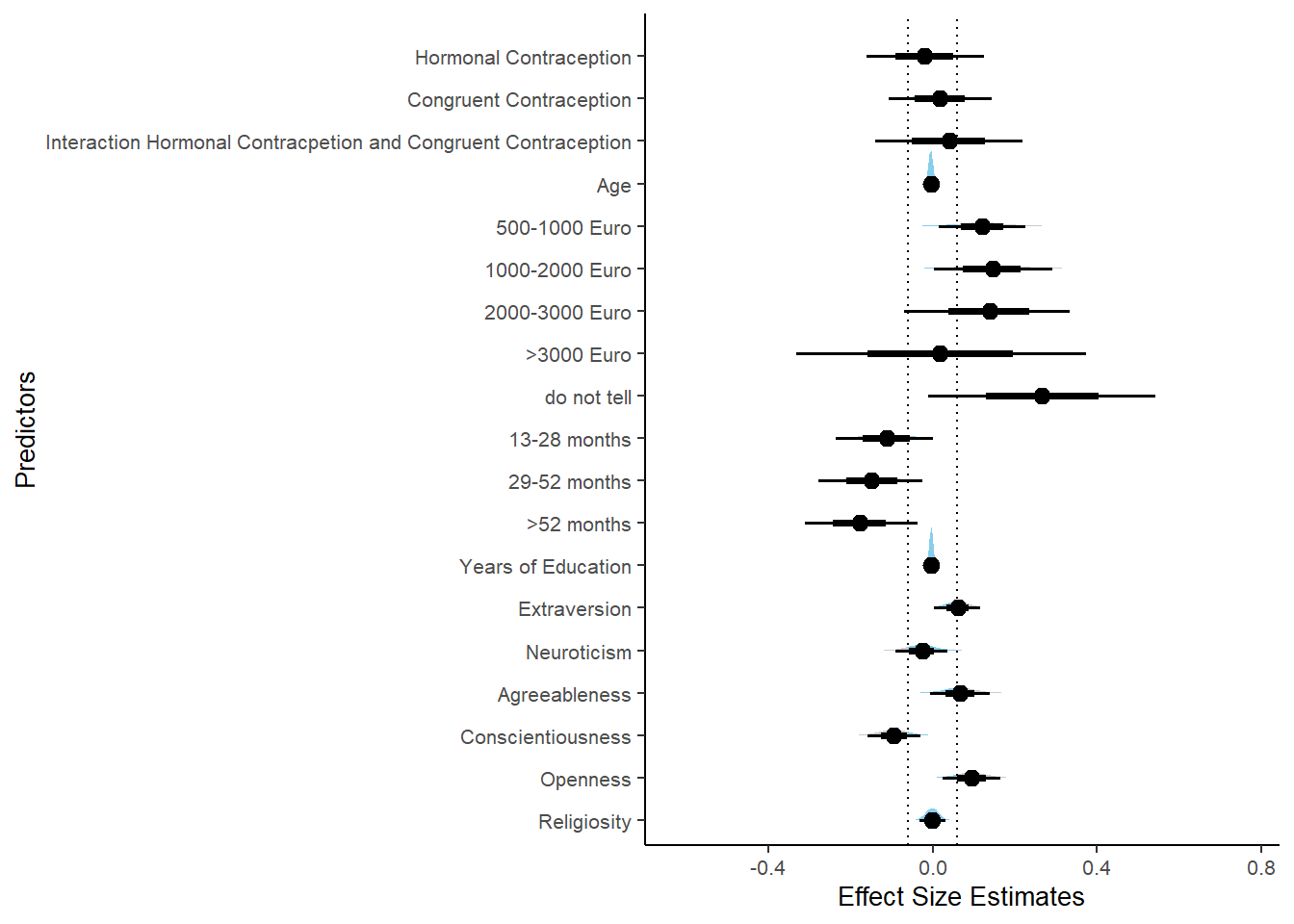
Sexual Frequency (Penetrative Intercourse)
Model
m_congruency_sexfreqpen_controlled = brm(diary_sex_active_sex_sum ~
offset(log(number_of_days)) +
contraception_hormonal * congruent_contraception +
age + net_income + relationship_duration_factor +
education_years +
bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open +
religiosity,
data = data, family = poisson(),
file = "m_congruency_sexfreqpen_controlled")## Warning: Rows containing NAs were excluded from the model.## Compiling Stan program...## recompiling to avoid crashing R session## Start sampling##
## SAMPLING FOR MODEL 'c63cc6f90adde557ca52c07be9ae7367' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 0 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 91.036 seconds (Warm-up)
## Chain 1: 115.326 seconds (Sampling)
## Chain 1: 206.362 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL 'c63cc6f90adde557ca52c07be9ae7367' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 0 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 96.757 seconds (Warm-up)
## Chain 2: 114.862 seconds (Sampling)
## Chain 2: 211.619 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL 'c63cc6f90adde557ca52c07be9ae7367' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 0 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 94.349 seconds (Warm-up)
## Chain 3: 115.023 seconds (Sampling)
## Chain 3: 209.372 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL 'c63cc6f90adde557ca52c07be9ae7367' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 0 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
## Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
## Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
## Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
## Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
## Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
## Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
## Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 94.231 seconds (Warm-up)
## Chain 4: 114.17 seconds (Sampling)
## Chain 4: 208.401 seconds (Total)
## Chain 4:## Warning: There were 4000 transitions after warmup that exceeded the maximum treedepth. Increase max_treedepth above 10. See
## http://mc-stan.org/misc/warnings.html#maximum-treedepth-exceeded## Warning: Examine the pairs() plot to diagnose sampling problems## Warning: The largest R-hat is 2.87, indicating chains have not mixed.
## Running the chains for more iterations may help. See
## http://mc-stan.org/misc/warnings.html#r-hat## Warning: Bulk Effective Samples Size (ESS) is too low, indicating posterior means and medians may be unreliable.
## Running the chains for more iterations may help. See
## http://mc-stan.org/misc/warnings.html#bulk-ess## Warning: Tail Effective Samples Size (ESS) is too low, indicating posterior variances and tail quantiles may be unreliable.
## Running the chains for more iterations may help. See
## http://mc-stan.org/misc/warnings.html#tail-essSummary
## Warning: Parts of the model have not converged (some Rhats are > 1.05). Be careful when analysing the results!
## We recommend running more iterations and/or setting stronger priors.## Family: poisson
## Links: mu = log
## Formula: diary_sex_active_sex_sum ~ offset(log(number_of_days)) + contraception_hormonal * congruent_contraception + age + net_income + relationship_duration_factor + education_years + bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open + religiosity
## Data: data (Number of observations: 622)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept 40.61 230.38 -280.16 358.71 2.87 5
## contraception_hormonalyes 0.14 0.05 0.06 0.22 1.14 39
## congruent_contraception1 -0.05 0.04 -0.11 0.01 1.08 40
## age -0.00 0.00 -0.01 0.00 1.06 52
## net_incomeeuro_500_1000 0.14 0.03 0.08 0.20 1.11 34
## net_incomeeuro_1000_2000 0.17 0.05 0.09 0.24 1.08 39
## net_incomeeuro_2000_3000 0.37 0.06 0.26 0.47 1.07 34
## net_incomeeuro_gt_3000 0.04 0.11 -0.16 0.19 1.14 29
## net_incomedont_tell 0.37 0.09 0.24 0.51 1.13 22
## relationship_duration_factorPartnered_upto12months -42.29 230.37 -360.31 278.53 2.87 5
## relationship_duration_factorPartnered_upto28months -42.42 230.37 -360.42 278.38 2.87 5
## relationship_duration_factorPartnered_upto52months -42.68 230.37 -360.71 278.12 2.87 5
## relationship_duration_factorPartnered_morethan52months -42.70 230.37 -360.69 278.13 2.87 5
## education_years -0.01 0.00 -0.01 -0.01 1.02 106
## bfi_extra -0.02 0.02 -0.05 0.01 1.14 26
## bfi_neuro -0.02 0.02 -0.05 0.01 1.19 19
## bfi_agree 0.09 0.02 0.06 0.13 1.05 47
## bfi_consc -0.03 0.02 -0.07 -0.00 1.24 14
## bfi_open 0.03 0.02 -0.00 0.07 1.08 40
## religiosity -0.01 0.01 -0.02 0.01 1.06 52
## contraception_hormonalyes:congruent_contraception1 0.05 0.06 -0.05 0.13 1.12 36
## Tail_ESS
## Intercept NA
## contraception_hormonalyes NA
## congruent_contraception1 NA
## age NA
## net_incomeeuro_500_1000 NA
## net_incomeeuro_1000_2000 NA
## net_incomeeuro_2000_3000 NA
## net_incomeeuro_gt_3000 NA
## net_incomedont_tell NA
## relationship_duration_factorPartnered_upto12months NA
## relationship_duration_factorPartnered_upto28months NA
## relationship_duration_factorPartnered_upto52months NA
## relationship_duration_factorPartnered_morethan52months NA
## education_years NA
## bfi_extra NA
## bfi_neuro NA
## bfi_agree NA
## bfi_consc NA
## bfi_open NA
## religiosity NA
## contraception_hormonalyes:congruent_contraception1 NA
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_congruency_sexfreqpen_controlled, range = c(-0.05, 0.05),
ci = 0.90,
parameters = "contraception"))## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.79), b_contraception_hormonalyes:congruent_contraception1 and b_congruent_contraception1 (r = 0.73). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## Picking joint bandwidth of 0.00659## Warning: Removed 1197 rows containing non-finite values (stat_density_ridges).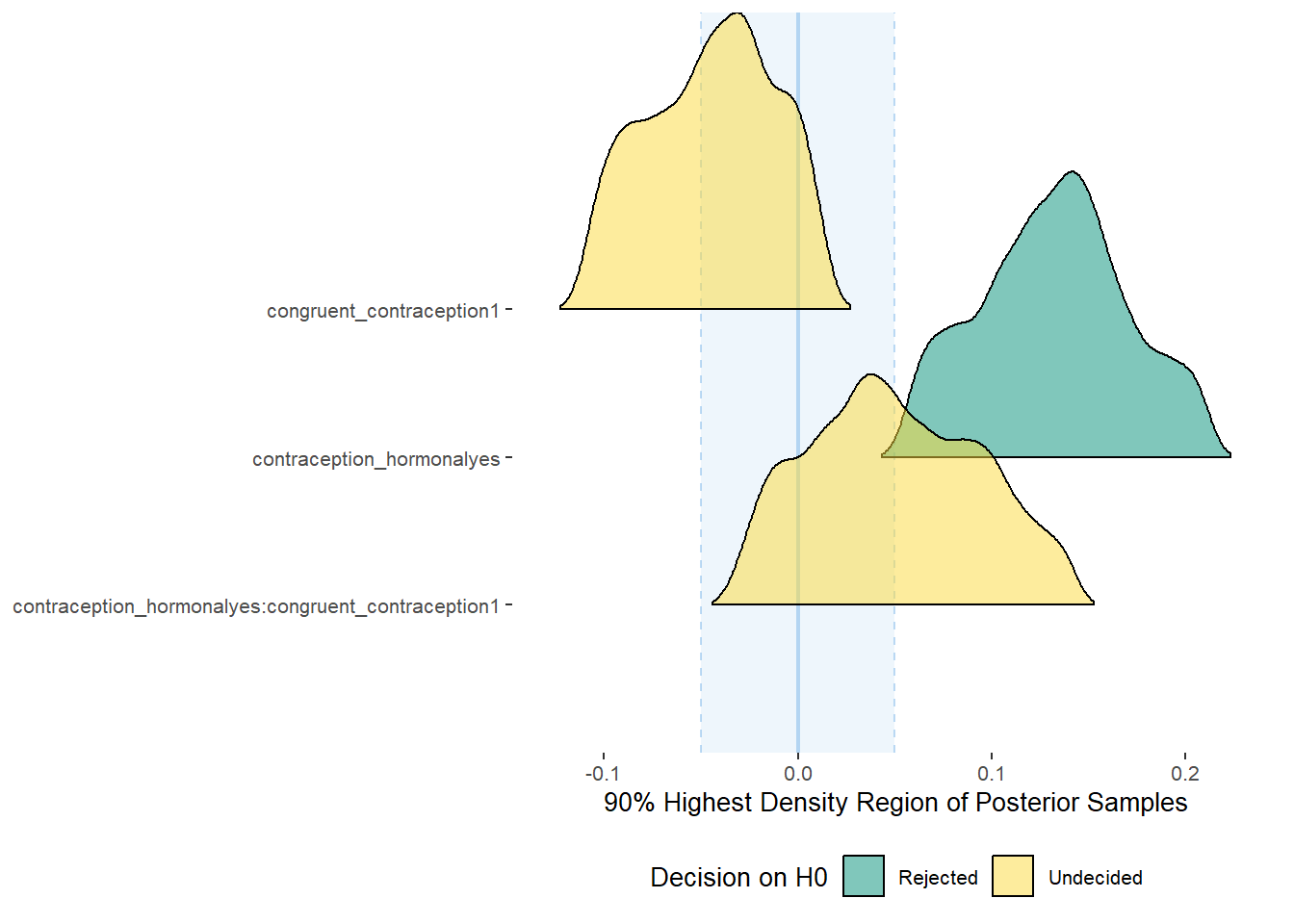
equivalence_test(m_congruency_sexfreqpen_controlled, range = c(-0.05, 0.05), ci = 0.90,
parameters = "contraception")## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.79), b_contraception_hormonalyes:congruent_contraception1 and b_congruent_contraception1 (r = 0.73). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## # A tibble: 3 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.05 0.05 0 Rejected 0.0556 0.211 fixed conditio~
## 2 b_congruent_co~ 0.9 -0.05 0.05 0.581 Undecided -0.110 0.0144 fixed conditio~
## 3 b_contraceptio~ 0.9 -0.05 0.05 0.524 Undecided -0.0349 0.142 fixed conditio~Plots
conditional_effects(m_congruency_sexfreqpen_controlled,
effects = "contraception_hormonal:congruent_contraception",
conditions = data.frame(number_of_days = 1))
Forest Plot for Effect Sizes
m_congruency_sexfreqpen_controlled %>%
spread_draws(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity) %>%
pivot_longer(cols = c(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_relationship_duration_factor",
"Relationship Duration",
ifelse(condition %contains% "b_net_income",
"Income",
NA)),
group = ifelse(condition %contains% "ontraception",
"Contraception", group),
condition = ifelse(condition == "b_contraception_hormonalyes",
"Hormonal Contraception",
ifelse(condition == "b_congruent_contraception1",
"Congruent Contraception",
ifelse(condition == "b_contraception_hormonalyes:congruent_contraception1",
"Interaction Hormonal Contracpetion and Congruent Contraception",
condition))),
condition = ifelse(condition == "b_age", "Age",
ifelse(condition == "b_net_incomeeuro_500_1000", "500-1000 Euro",
ifelse(condition == "b_net_incomeeuro_1000_2000", "1000-2000 Euro",
ifelse(condition == "b_net_incomeeuro_2000_3000", "2000-3000 Euro",
ifelse(condition == "b_net_incomeeuro_gt_3000", ">3000 Euro",
ifelse(condition == "b_net_incomedont_tell", "do not tell",
ifelse(condition == "b_relationship_duration_factorPartnered_upto28months",
"13-28 months",
ifelse(condition == "b_relationship_duration_factorPartnered_upto52months",
"29-52 months",
ifelse(condition == "b_relationship_duration_factorPartnered_morethan52months",
">52 months",
ifelse(condition == "b_education_years", "Years of Education",
ifelse(condition == "b_bfi_extra", "Extraversion",
ifelse(condition == "b_bfi_neuro", "Neuroticism",
ifelse(condition == "b_bfi_agree", "Agreeableness",
ifelse(condition == "b_bfi_consc", "Conscientiousness",
ifelse(condition == "b_bfi_open", "Openness",
ifelse(condition == "b_religiosity", "Religiosity",
condition)))))))))))))))),
group = ifelse(is.na(group), condition, group),
condition = factor(condition, levels = rev(c("Hormonal Contraception",
"Congruent Contraception",
"Interaction Hormonal Contracpetion and Congruent Contraception",
"Age",
"500-1000 Euro", "1000-2000 Euro",
"2000-3000 Euro", ">3000 Euro", "do not tell",
"13-28 months", "29-52 months",
">52 months",
"Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))),
group = factor(group, levels = c("Contraception", "Age", "Income",
"Relationship Duration","Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.05))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.05, 0.05), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")
Masturbation Frequency
Model
m_congruency_masfreq_controlled = brm(diary_masturbation_sum ~
offset(log(number_of_days)) +
contraception_hormonal * congruent_contraception +
age + net_income + relationship_duration_factor +
education_years +
bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open +
religiosity,
data = data, family = poisson(),
file = "m_congruency_masfreq_controlled")Summary
## Family: poisson
## Links: mu = log
## Formula: diary_masturbation_sum ~ offset(log(number_of_days)) + contraception_hormonal * congruent_contraception + age + net_income + relationship_duration_factor + education_years + bfi_extra + bfi_neuro + bfi_agree + bfi_consc + bfi_open + religiosity
## Data: data (Number of observations: 622)
## Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup draws = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-90% CI u-90% CI Rhat Bulk_ESS
## Intercept -1.74 0.23 -2.12 -1.36 1.00 4718
## contraception_hormonalyes -0.38 0.06 -0.47 -0.28 1.00 3090
## congruent_contraception1 0.04 0.05 -0.04 0.12 1.00 3121
## age -0.01 0.00 -0.02 -0.00 1.00 4213
## net_incomeeuro_500_1000 0.18 0.04 0.11 0.25 1.00 3789
## net_incomeeuro_1000_2000 0.21 0.06 0.12 0.31 1.00 3356
## net_incomeeuro_2000_3000 0.07 0.08 -0.07 0.20 1.00 3447
## net_incomeeuro_gt_3000 -0.13 0.15 -0.37 0.11 1.00 4879
## net_incomedont_tell -0.18 0.12 -0.37 0.01 1.00 4786
## relationship_duration_factorPartnered_upto28months -0.03 0.04 -0.11 0.04 1.00 3792
## relationship_duration_factorPartnered_upto52months -0.07 0.05 -0.15 0.01 1.00 3586
## relationship_duration_factorPartnered_morethan52months -0.20 0.05 -0.28 -0.12 1.00 3790
## education_years 0.00 0.00 -0.00 0.01 1.00 5824
## bfi_extra -0.02 0.02 -0.06 0.02 1.00 4565
## bfi_neuro 0.00 0.02 -0.04 0.04 1.00 4670
## bfi_agree 0.01 0.03 -0.04 0.06 1.00 4377
## bfi_consc -0.22 0.03 -0.26 -0.18 1.00 5098
## bfi_open 0.20 0.03 0.15 0.25 1.00 5236
## religiosity -0.06 0.01 -0.08 -0.04 1.00 4483
## contraception_hormonalyes:congruent_contraception1 0.10 0.07 -0.02 0.22 1.00 2862
## Tail_ESS
## Intercept 3203
## contraception_hormonalyes 2694
## congruent_contraception1 2405
## age 3421
## net_incomeeuro_500_1000 3027
## net_incomeeuro_1000_2000 2833
## net_incomeeuro_2000_3000 3065
## net_incomeeuro_gt_3000 3343
## net_incomedont_tell 3204
## relationship_duration_factorPartnered_upto28months 3010
## relationship_duration_factorPartnered_upto52months 3355
## relationship_duration_factorPartnered_morethan52months 3275
## education_years 3022
## bfi_extra 2643
## bfi_neuro 2568
## bfi_agree 3140
## bfi_consc 3185
## bfi_open 3192
## religiosity 3143
## contraception_hormonalyes:congruent_contraception1 2727
##
## Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Comparison with ROPE
plot(equivalence_test(m_congruency_masfreq_controlled, range = c(-0.05, 0.05), ci = 0.90,
parameters = "contraception"))## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.8). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## Picking joint bandwidth of 0.00819## Warning: Removed 1197 rows containing non-finite values (stat_density_ridges).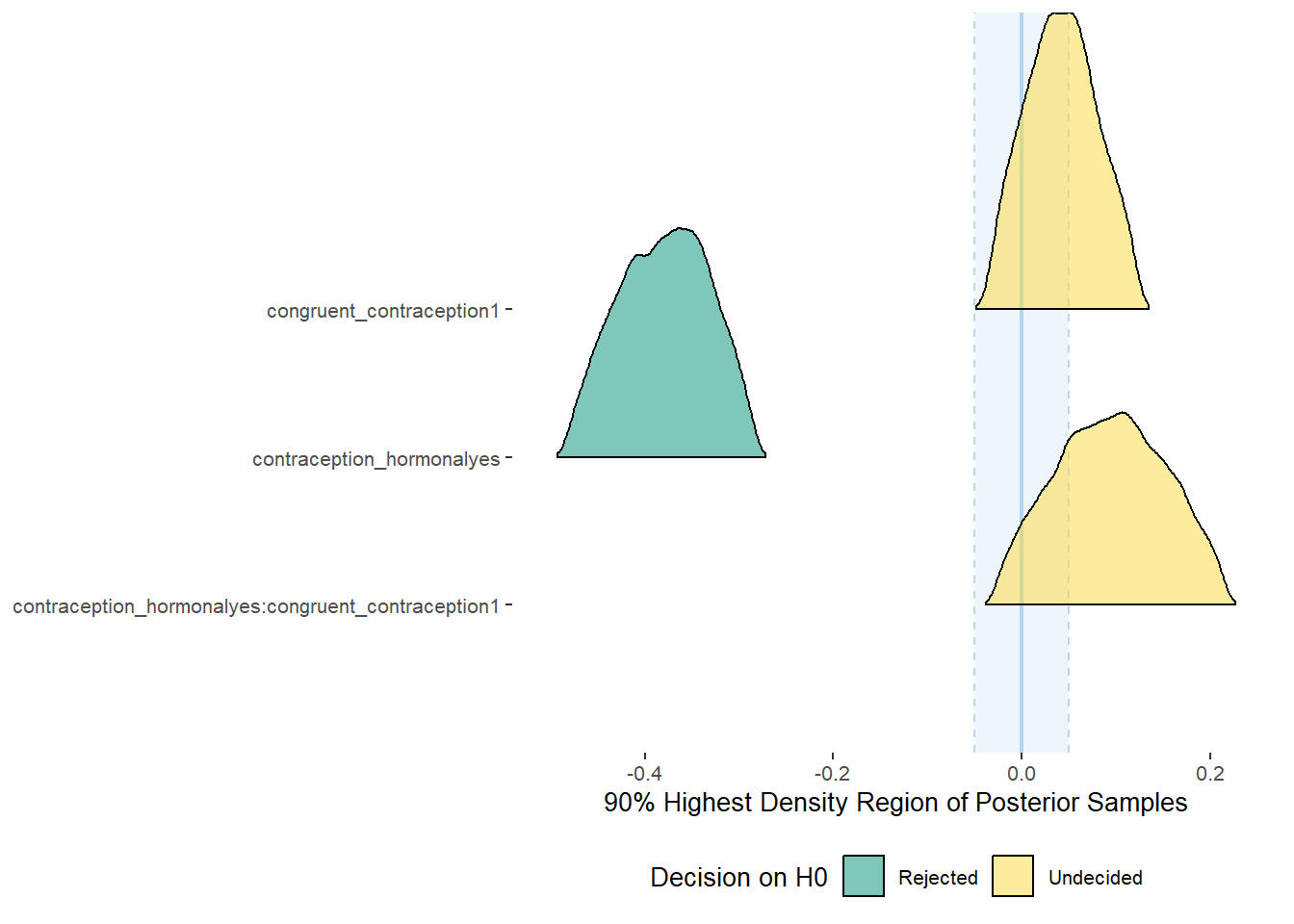
equivalence_test(m_congruency_masfreq_controlled, range = c(-0.05, 0.05), ci = 0.90,
parameters = "contraception")## Possible multicollinearity between b_contraception_hormonalyes:congruent_contraception1 and b_contraception_hormonalyes (r = 0.8). This might lead to inappropriate results. See 'Details' in '?equivalence_test'.## # A tibble: 3 x 10
## Parameter CI ROPE_low ROPE_high ROPE_Percentage ROPE_Equivalence HDI_low HDI_high Effects Component
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 b_contraceptio~ 0.9 -0.05 0.05 0 Rejected -0.479 -0.286 fixed conditio~
## 2 b_congruent_co~ 0.9 -0.05 0.05 0.573 Undecided -0.0332 0.121 fixed conditio~
## 3 b_contraceptio~ 0.9 -0.05 0.05 0.244 Undecided -0.0244 0.213 fixed conditio~Plots
conditional_effects(m_congruency_masfreq_controlled,
effects = "contraception_hormonal:congruent_contraception",
conditions = data.frame(number_of_days = 1))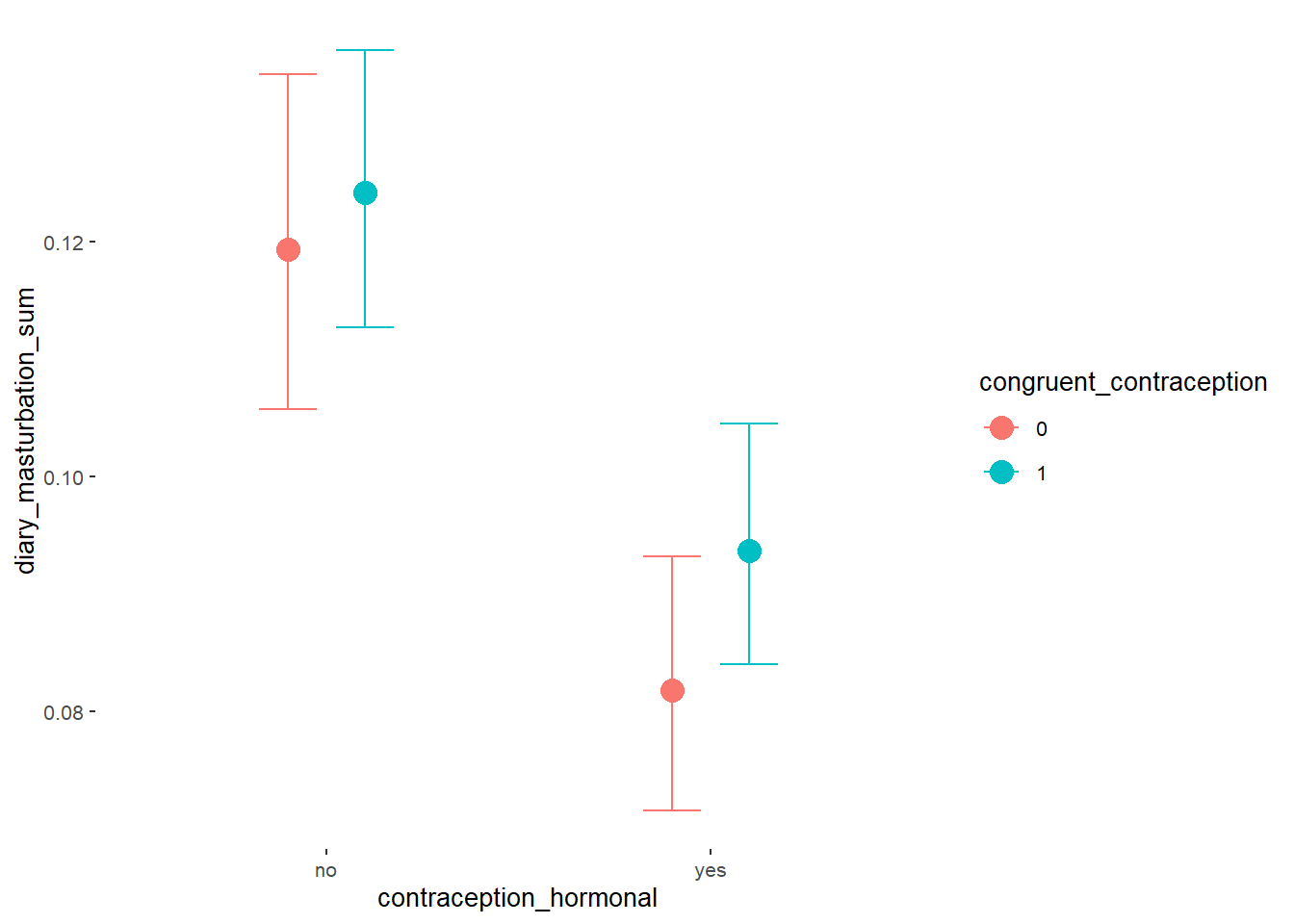
Forest Plot for Effect Sizes
m_congruency_masfreq_controlled %>%
spread_draws(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity) %>%
pivot_longer(cols = c(b_contraception_hormonalyes, b_congruent_contraception1,
`b_contraception_hormonalyes:congruent_contraception1`,
b_age,
b_net_incomeeuro_500_1000, b_net_incomeeuro_1000_2000,
b_net_incomeeuro_2000_3000, b_net_incomeeuro_gt_3000, b_net_incomedont_tell,
b_relationship_duration_factorPartnered_upto28months,
b_relationship_duration_factorPartnered_upto52months,
b_relationship_duration_factorPartnered_morethan52months,
b_education_years,
b_bfi_extra, b_bfi_neuro, b_bfi_agree, b_bfi_consc, b_bfi_open,
b_religiosity),
names_to = "condition",
values_to = "r_condition") %>%
mutate(condition_mean = r_condition,
group = ifelse(condition %contains% "b_relationship_duration_factor",
"Relationship Duration",
ifelse(condition %contains% "b_net_income",
"Income",
NA)),
group = ifelse(condition %contains% "ontraception",
"Contraception", group),
condition = ifelse(condition == "b_contraception_hormonalyes",
"Hormonal Contraception",
ifelse(condition == "b_congruent_contraception1",
"Congruent Contraception",
ifelse(condition == "b_contraception_hormonalyes:congruent_contraception1",
"Interaction Hormonal Contracpetion and Congruent Contraception",
condition))),
condition = ifelse(condition == "b_age", "Age",
ifelse(condition == "b_net_incomeeuro_500_1000", "500-1000 Euro",
ifelse(condition == "b_net_incomeeuro_1000_2000", "1000-2000 Euro",
ifelse(condition == "b_net_incomeeuro_2000_3000", "2000-3000 Euro",
ifelse(condition == "b_net_incomeeuro_gt_3000", ">3000 Euro",
ifelse(condition == "b_net_incomedont_tell", "do not tell",
ifelse(condition == "b_relationship_duration_factorPartnered_upto28months",
"13-28 months",
ifelse(condition == "b_relationship_duration_factorPartnered_upto52months",
"29-52 months",
ifelse(condition == "b_relationship_duration_factorPartnered_morethan52months",
">52 months",
ifelse(condition == "b_education_years", "Years of Education",
ifelse(condition == "b_bfi_extra", "Extraversion",
ifelse(condition == "b_bfi_neuro", "Neuroticism",
ifelse(condition == "b_bfi_agree", "Agreeableness",
ifelse(condition == "b_bfi_consc", "Conscientiousness",
ifelse(condition == "b_bfi_open", "Openness",
ifelse(condition == "b_religiosity", "Religiosity",
condition)))))))))))))))),
group = ifelse(is.na(group), condition, group),
condition = factor(condition, levels = rev(c("Hormonal Contraception",
"Congruent Contraception",
"Interaction Hormonal Contracpetion and Congruent Contraception",
"Age",
"500-1000 Euro", "1000-2000 Euro",
"2000-3000 Euro", ">3000 Euro", "do not tell",
"13-28 months", "29-52 months",
">52 months",
"Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))),
group = factor(group, levels = c("Contraception", "Age", "Income",
"Relationship Duration","Years of Education",
"Extraversion", "Neuroticism", "Agreeableness",
"Conscientiousness","Openness","Religiosity"))) %>%
ggplot(aes(y = condition,
x = condition_mean,
fill = stat(abs(x) < 0.05))) +
stat_halfeye() +
geom_vline(xintercept = c(-0.05, 0.05), linetype = "dotted") +
apatheme +
theme(legend.position = "none") +
scale_fill_manual(values = c("gray80", "skyblue")) +
labs(x = "Effect Size Estimates", y = "Predictors")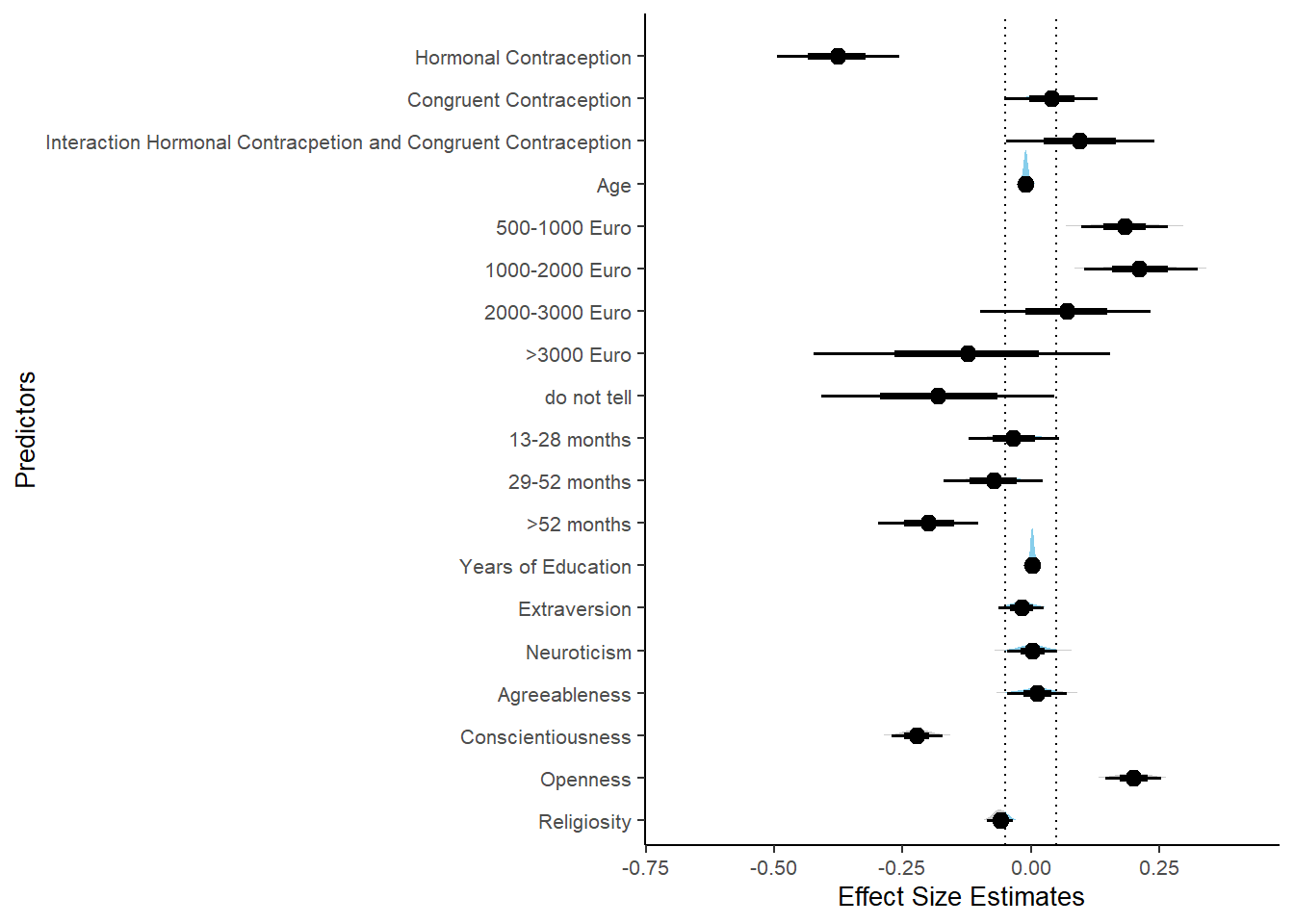
Model Comparison
Attractiveness of partner
LooIC first model: 1726.1
LooIC second model: 1726.72
Model Comparisons: The difference between models is -0.31 compared to a standard error of 1.86
## Output of model 'm_hc_atrr':
##
## Computed from 4000 by 774 log-likelihood matrix
##
## Estimate SE
## elpd_loo -863.0 23.1
## p_loo 3.3 0.4
## looic 1726.1 46.2
## ------
## Monte Carlo SE of elpd_loo is 0.0.
##
## All Pareto k estimates are good (k < 0.5).
## See help('pareto-k-diagnostic') for details.
##
## Output of model 'm_congruency_atrr':
##
## Computed from 4000 by 774 log-likelihood matrix
##
## Estimate SE
## elpd_loo -863.4 23.2
## p_loo 5.3 0.5
## looic 1726.7 46.5
## ------
## Monte Carlo SE of elpd_loo is 0.0.
##
## All Pareto k estimates are good (k < 0.5).
## See help('pareto-k-diagnostic') for details.
##
## Model comparisons:
## elpd_diff se_diff
## m_hc_atrr 0.0 0.0
## m_congruency_atrr -0.3 1.9Relationship satisfaction
LooIC first model: 872.85
LooIC second model: 870.03
Model Comparisons: The difference between models is -1.41 compared to a standard error of 2.66
## Output of model 'm_hc_relsat':
##
## Computed from 4000 by 774 log-likelihood matrix
##
## Estimate SE
## elpd_loo -436.4 26.6
## p_loo 3.9 0.5
## looic 872.9 53.2
## ------
## Monte Carlo SE of elpd_loo is 0.0.
##
## All Pareto k estimates are good (k < 0.5).
## See help('pareto-k-diagnostic') for details.
##
## Output of model 'm_congruency_relsat':
##
## Computed from 4000 by 774 log-likelihood matrix
##
## Estimate SE
## elpd_loo -435.0 26.3
## p_loo 5.8 0.6
## looic 870.0 52.5
## ------
## Monte Carlo SE of elpd_loo is 0.0.
##
## All Pareto k estimates are good (k < 0.5).
## See help('pareto-k-diagnostic') for details.
##
## Model comparisons:
## elpd_diff se_diff
## m_congruency_relsat 0.0 0.0
## m_hc_relsat -1.4 2.7Sexual Satisfaction
m_hc_sexsat$data$satisfaction_sexual_intercourse =
as.numeric(m_hc_sexsat$data$satisfaction_sexual_intercourse)
m_congruency_sexsat$data$satisfaction_sexual_intercourse =
as.numeric(m_congruency_sexsat$data$satisfaction_sexual_intercourse)
compare_models = loo(m_hc_sexsat, m_congruency_sexsat)LooIC first model: 2276.46
LooIC second model: 2277.93
Model Comparisons: The difference between models is -0.74 compared to a standard error of 1.61
## Output of model 'm_hc_sexsat':
##
## Computed from 4000 by 774 log-likelihood matrix
##
## Estimate SE
## elpd_loo -1138.2 21.0
## p_loo 3.2 0.3
## looic 2276.5 42.0
## ------
## Monte Carlo SE of elpd_loo is 0.0.
##
## All Pareto k estimates are good (k < 0.5).
## See help('pareto-k-diagnostic') for details.
##
## Output of model 'm_congruency_sexsat':
##
## Computed from 4000 by 774 log-likelihood matrix
##
## Estimate SE
## elpd_loo -1139.0 21.0
## p_loo 5.1 0.4
## looic 2277.9 42.1
## ------
## Monte Carlo SE of elpd_loo is 0.0.
##
## All Pareto k estimates are good (k < 0.5).
## See help('pareto-k-diagnostic') for details.
##
## Model comparisons:
## elpd_diff se_diff
## m_hc_sexsat 0.0 0.0
## m_congruency_sexsat -0.7 1.6LS0tDQp0aXRsZTogIkFuYWx5c2VzIEVmZmVjdHMgb2YgQ29udHJhY2VwdGlvbiINCm91dHB1dDoNCiAgaHRtbF9kb2N1bWVudDoNCiAgICB0b2M6IHRydWUNCiAgICB0b2NfZGVwdGg6IDQNCiAgICB0b2NfZmxvYXQ6IHRydWUNCiAgICBjb2RlX2ZvbGRpbmc6ICdoaWRlJw0KICAgIHNlbGZfY29udGFpbmVkOiBmYWxzZQ0KZWRpdG9yX29wdGlvbnM6IA0KICBjaHVua19vdXRwdXRfdHlwZTogY29uc29sZQ0KLS0tDQogIA0KICANCiMjIERhdGEgDQpgYGB7ciByZXN1bHRzPSdoaWRlJyxtZXNzYWdlPUYsd2FybmluZz1GfQ0Kc291cmNlKCIwX2hlbHBlcnMuUiIpDQoNCmxvYWQoImRhdGEvY2xlYW5lZF9zZWxlY3RlZF93cmFuZ2xlZC5yZGF0YSIpDQpgYGANCg0KDQojIyBFZmZlY3RzIG9mIEhvcm1vbmFsIENvbnRyYWNlcHRpdmVzIHsudGFic2V0fQ0KIyMjIEF0dHJhY3RpdmVuZXNzIG9mIFBhcnRuZXIgey50YWJzZXR9DQojIyMjIE1vZGVsDQpgYGB7cn0NCm1faGNfYXRyciA9IGJybShhdHRyYWN0aXZlbmVzc19wYXJ0bmVyIH4gY29udHJhY2VwdGlvbl9ob3Jtb25hbCwNCiAgICAgICAgICAgICAgICBkYXRhID0gZGF0YSwgZmFtaWx5ID0gZ2F1c3NpYW4oKSwNCiAgICAgICAgICAgICAgICBmaWxlID0gIm1faGNfYXRyciIpDQpgYGANCg0KIyMjIyBTdW1tYXJ5DQpgYGB7cn0NCnN1bW1hcnkobV9oY19hdHJyLCBpbnRlcnZhbHMgPSBULCBwcm9iID0gMC45MCkNCmBgYA0KDQojIyMjIENvbXBhcmlzb24gd2l0aCBST1BFDQpgYGB7cn0NCnBsb3QoZXF1aXZhbGVuY2VfdGVzdChtX2hjX2F0cnIsIHJhbmdlID0gYygtMC4wNywgMC4wNyksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKSkNCmVxdWl2YWxlbmNlX3Rlc3QobV9oY19hdHJyLCByYW5nZSA9IGMoLTAuMDcsIDAuMDcpLCBjaSA9IDAuOTAsDQogICAgICAgICAgICAgICAgICAgICAgcGFyYW1ldGVycyA9ICJjb250cmFjZXB0aW9uIikNCmBgYA0KDQojIyMjIFBsb3RzDQpgYGB7ciB3YXJuaW5nID0gRkFMU0V9DQpjb25kaXRpb25hbF9lZmZlY3RzKG1faGNfYXRyciwgYXNrID0gRkFMU0UpDQpgYGANCg0KIyMjIyBGb3Jlc3QgUGxvdCBmb3IgRWZmZWN0IFNpemVzIHsuYWN0aXZlfQ0KYGBge3J9DQptX2hjX2F0cnIgJT4lDQogIHNwcmVhZF9kcmF3cyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMpICU+JQ0KICBwaXZvdF9sb25nZXIoY29scyA9IGMoYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzKSwNCiAgICAgICAgICAgICAgIG5hbWVzX3RvID0gImNvbmRpdGlvbiIsDQogICAgICAgICAgICAgICB2YWx1ZXNfdG8gPSAicl9jb25kaXRpb24iKSAlPiUNCiAgbXV0YXRlKGNvbmRpdGlvbl9tZWFuID0gcl9jb25kaXRpb24sDQogICAgICAgICBncm91cCA9IGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAiYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJDb250cmFjZXB0aW9uIiwgTkEpLA0KICAgICAgICAgY29uZGl0aW9uID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLCBOQSkpICU+JQ0KICBnZ3Bsb3QoYWVzKHkgPSBjb25kaXRpb24sDQogICAgICAgICAgICAgeCA9IGNvbmRpdGlvbl9tZWFuLA0KICAgICAgICAgICAgIGZpbGwgPSBzdGF0KGFicyh4KSA8IDAuMDcpKSkgKw0KICBzdGF0X2hhbGZleWUoKSArDQogIGdlb21fdmxpbmUoeGludGVyY2VwdCA9IGMoLTAuMDcsIDAuMDcpLCBsaW5ldHlwZSA9ICJkb3R0ZWQiKSArDQogIGFwYXRoZW1lICsNCiAgdGhlbWUobGVnZW5kLnBvc2l0aW9uID0gIm5vbmUiKSArDQogIHNjYWxlX2ZpbGxfbWFudWFsKHZhbHVlcyA9IGMoImdyYXk4MCIsICJza3libHVlIikpICsNCiAgbGFicyh4ID0gIkVmZmVjdCBTaXplIEVzdGltYXRlcyIsIHkgPSAiUHJlZGljdG9ycyIpICsNCiAgeGxpbSAoLTAuNiwgMC42KQ0KYGBgDQoNCiMjIyBSZWxhdGlvbnNoaXAgU2F0aXNmYWN0aW9uIHsudGFic2V0fQ0KIyMjIyBNb2RlbA0KYGBge3J9DQptX2hjX3JlbHNhdCA9IGJybShyZWxhdGlvbnNoaXBfc2F0aXNmYWN0aW9uIH4gY29udHJhY2VwdGlvbl9ob3Jtb25hbCwNCiAgICAgICAgICAgICAgICBkYXRhID0gZGF0YSwgZmFtaWx5ID0gZ2F1c3NpYW4oKSwNCiAgICAgICAgICAgICAgICBmaWxlID0gIm1faGNfcmVsc2F0IikNCmBgYA0KDQojIyMjIFN1bW1hcnkNCmBgYHtyfQ0Kc3VtbWFyeShtX2hjX3JlbHNhdCwgaW50ZXJ2YWxzID0gVCwgcHJvYiA9IDAuOTApDQpgYGANCg0KIyMjIyBDb21wYXJpc29uIHdpdGggUk9QRQ0KYGBge3J9DQpwbG90KGVxdWl2YWxlbmNlX3Rlc3QobV9oY19yZWxzYXQsIHJhbmdlID0gYygtMC4wNCwgMC4wNCksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKSkNCmVxdWl2YWxlbmNlX3Rlc3QobV9oY19yZWxzYXQsIHJhbmdlID0gYygtMC4wNCwgMC4wNCksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKQ0KYGBgDQoNCiMjIyMgUGxvdHMNCmBgYHtyIHdhcm5pbmcgPSBGQUxTRX0NCmNvbmRpdGlvbmFsX2VmZmVjdHMobV9oY19yZWxzYXQsIGFzayA9IEZBTFNFKQ0KYGBgDQoNCiMjIyMgRm9yZXN0IFBsb3QgZm9yIEVmZmVjdCBTaXplcyB7LmFjdGl2ZX0NCmBgYHtyfQ0KbV9oY19yZWxzYXQgJT4lDQogIHNwcmVhZF9kcmF3cyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMpICU+JQ0KICBwaXZvdF9sb25nZXIoY29scyA9IGMoYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzKSwNCiAgICAgICAgICAgICAgIG5hbWVzX3RvID0gImNvbmRpdGlvbiIsDQogICAgICAgICAgICAgICB2YWx1ZXNfdG8gPSAicl9jb25kaXRpb24iKSAlPiUNCiAgbXV0YXRlKGNvbmRpdGlvbl9tZWFuID0gcl9jb25kaXRpb24sDQogICAgICAgICBncm91cCA9IGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAiYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJDb250cmFjZXB0aW9uIiwgTkEpLA0KICAgICAgICAgY29uZGl0aW9uID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLCBOQSkpICU+JQ0KICBnZ3Bsb3QoYWVzKHkgPSBjb25kaXRpb24sDQogICAgICAgICAgICAgeCA9IGNvbmRpdGlvbl9tZWFuLA0KICAgICAgICAgICAgIGZpbGwgPSBzdGF0KGFicyh4KSA8IDAuMDQpKSkgKw0KICBzdGF0X2hhbGZleWUoKSArDQogIGdlb21fdmxpbmUoeGludGVyY2VwdCA9IGMoLTAuMDQsIDAuMDQpLCBsaW5ldHlwZSA9ICJkb3R0ZWQiKSArDQogIGFwYXRoZW1lICsNCiAgdGhlbWUobGVnZW5kLnBvc2l0aW9uID0gIm5vbmUiKSArDQogIHNjYWxlX2ZpbGxfbWFudWFsKHZhbHVlcyA9IGMoImdyYXk4MCIsICJza3libHVlIikpICsNCiAgbGFicyh4ID0gIkVmZmVjdCBTaXplIEVzdGltYXRlcyIsIHkgPSAiUHJlZGljdG9ycyIpICsNCiAgeGxpbSAoLTAuNiwgMC42KQ0KYGBgDQoNCg0KIyMjIFNleHVhbCBTYXRpc2ZhY3Rpb24gey50YWJzZXR9DQojIyMjIE1vZGVsDQpgYGB7cn0NCm1faGNfc2V4c2F0ID0gYnJtKHNhdGlzZmFjdGlvbl9zZXh1YWxfaW50ZXJjb3Vyc2UgfiBjb250cmFjZXB0aW9uX2hvcm1vbmFsLA0KICAgICAgICAgICAgICAgIGRhdGEgPSBkYXRhLCBmYW1pbHkgPSBnYXVzc2lhbigpLA0KICAgICAgICAgICAgICAgIGZpbGUgPSAibV9oY19zZXhzYXQiKQ0KYGBgDQoNCiMjIyMgU3VtbWFyeQ0KYGBge3J9DQpzdW1tYXJ5KG1faGNfc2V4c2F0LCBpbnRlcnZhbHMgPSBULCBwcm9iID0gMC45MCkNCmBgYA0KDQojIyMjIENvbXBhcmlzb24gd2l0aCBST1BFDQpgYGB7cn0NCnBsb3QoZXF1aXZhbGVuY2VfdGVzdChtX2hjX3NleHNhdCwgcmFuZ2UgPSBjKC0wLjExLCAwLjExKSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpKQ0KZXF1aXZhbGVuY2VfdGVzdChtX2hjX3NleHNhdCwgcmFuZ2UgPSBjKC0wLjExLCAwLjExKSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpDQpgYGANCg0KIyMjIyBQbG90cw0KYGBge3Igd2FybmluZyA9IEZBTFNFfQ0KY29uZGl0aW9uYWxfZWZmZWN0cyhtX2hjX3NleHNhdCwgYXNrID0gRkFMU0UpDQpgYGANCg0KIyMjIyBGb3Jlc3QgUGxvdCBmb3IgRWZmZWN0IFNpemVzIHsuYWN0aXZlfQ0KYGBge3J9DQptX2hjX3NleHNhdCAlPiUNCiAgc3ByZWFkX2RyYXdzKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcykgJT4lDQogIHBpdm90X2xvbmdlcihjb2xzID0gYyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMpLA0KICAgICAgICAgICAgICAgbmFtZXNfdG8gPSAiY29uZGl0aW9uIiwNCiAgICAgICAgICAgICAgIHZhbHVlc190byA9ICJyX2NvbmRpdGlvbiIpICU+JQ0KICBtdXRhdGUoY29uZGl0aW9uX21lYW4gPSByX2NvbmRpdGlvbiwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnRyYWNlcHRpb24iLCBOQSksDQogICAgICAgICBjb25kaXRpb24gPSBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAiSG9ybW9uYWwgQ29udHJhY2VwdGlvbiIsIE5BKSkgJT4lDQogIGdncGxvdChhZXMoeSA9IGNvbmRpdGlvbiwNCiAgICAgICAgICAgICB4ID0gY29uZGl0aW9uX21lYW4sDQogICAgICAgICAgICAgZmlsbCA9IHN0YXQoYWJzKHgpIDwgMC4xMSkpKSArDQogIHN0YXRfaGFsZmV5ZSgpICsNCiAgZ2VvbV92bGluZSh4aW50ZXJjZXB0ID0gYygtMC4xMSwgMC4xMSksIGxpbmV0eXBlID0gImRvdHRlZCIpICsNCiAgYXBhdGhlbWUgKw0KICB0aGVtZShsZWdlbmQucG9zaXRpb24gPSAibm9uZSIpICsNCiAgc2NhbGVfZmlsbF9tYW51YWwodmFsdWVzID0gYygiZ3JheTgwIiwgInNreWJsdWUiKSkgKw0KICBsYWJzKHggPSAiRWZmZWN0IFNpemUgRXN0aW1hdGVzIiwgeSA9ICJQcmVkaWN0b3JzIikgKw0KICB4bGltICgtMC42LCAwLjYpDQpgYGANCg0KIyMjIExpYmlkbyB7LnRhYnNldH0NCg0KIyMjIyBNb2RlbA0KYGBge3J9DQptX2hjX2xpYmlkbyA9IGJybShkaWFyeV9saWJpZG9fbWVhbiB+IGNvbnRyYWNlcHRpb25faG9ybW9uYWwsDQogICAgICAgICAgICAgICAgZGF0YSA9IGRhdGEsIGZhbWlseSA9IGdhdXNzaWFuKCksDQogICAgICAgICAgICAgICAgZmlsZSA9ICJtX2hjX2xpYmlkbyIpDQpgYGANCg0KIyMjIyBTdW1tYXJ5DQpgYGB7cn0NCnN1bW1hcnkobV9oY19saWJpZG8sIGludGVydmFscyA9IFQsIHByb2IgPSAwLjkwKQ0KYGBgDQoNCiMjIyMgQ29tcGFyaXNvbiB3aXRoIFJPUEUNCmBgYHtyfQ0KcGxvdChlcXVpdmFsZW5jZV90ZXN0KG1faGNfbGliaWRvLCByYW5nZSA9IGMoLTAuMDYsIDAuMDYpLCBjaSA9IDAuOTAsDQogICAgICAgICAgICAgICAgICAgICAgcGFyYW1ldGVycyA9ICJjb250cmFjZXB0aW9uIikpDQplcXVpdmFsZW5jZV90ZXN0KG1faGNfbGliaWRvLCByYW5nZSA9IGMoLTAuMDYsIDAuMDYpLCBjaSA9IDAuOTAsDQogICAgICAgICAgICAgICAgICAgICAgcGFyYW1ldGVycyA9ICJjb250cmFjZXB0aW9uIikNCmBgYA0KDQojIyMjIFBsb3RzDQpgYGB7ciB3YXJuaW5nID0gRkFMU0V9DQpjb25kaXRpb25hbF9lZmZlY3RzKG1faGNfbGliaWRvLCBhc2sgPSBGQUxTRSkNCmBgYA0KDQojIyMjIEZvcmVzdCBQbG90IGZvciBFZmZlY3QgU2l6ZXMgey5hY3RpdmV9DQpgYGB7cn0NCm1faGNfbGliaWRvICU+JQ0KICBzcHJlYWRfZHJhd3MoYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzKSAlPiUNCiAgcGl2b3RfbG9uZ2VyKGNvbHMgPSBjKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcyksDQogICAgICAgICAgICAgICBuYW1lc190byA9ICJjb25kaXRpb24iLA0KICAgICAgICAgICAgICAgdmFsdWVzX3RvID0gInJfY29uZGl0aW9uIikgJT4lDQogIG11dGF0ZShjb25kaXRpb25fbWVhbiA9IHJfY29uZGl0aW9uLA0KICAgICAgICAgZ3JvdXAgPSBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAiQ29udHJhY2VwdGlvbiIsIE5BKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAiYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJIb3Jtb25hbCBDb250cmFjZXB0aW9uIiwgTkEpKSAlPiUNCiAgZ2dwbG90KGFlcyh5ID0gY29uZGl0aW9uLA0KICAgICAgICAgICAgIHggPSBjb25kaXRpb25fbWVhbiwNCiAgICAgICAgICAgICBmaWxsID0gc3RhdChhYnMoeCkgPCAwLjA2KSkpICsNCiAgc3RhdF9oYWxmZXllKCkgKw0KICBnZW9tX3ZsaW5lKHhpbnRlcmNlcHQgPSBjKC0wLjA2LCAwLjA2KSwgbGluZXR5cGUgPSAiZG90dGVkIikgKw0KICBhcGF0aGVtZSArDQogIHRoZW1lKGxlZ2VuZC5wb3NpdGlvbiA9ICJub25lIikgKw0KICBzY2FsZV9maWxsX21hbnVhbCh2YWx1ZXMgPSBjKCJncmF5ODAiLCAic2t5Ymx1ZSIpKSArDQogIGxhYnMoeCA9ICJFZmZlY3QgU2l6ZSBFc3RpbWF0ZXMiLCB5ID0gIlByZWRpY3RvcnMiKSArDQogIHhsaW0gKC0wLjYsIDAuNikNCmBgYA0KDQoNCiMjIyBTZXh1YWwgRnJlcXVlbmN5IChQZW5ldHJhdGl2ZSBJbnRlcmNvdXJzZSkgey50YWJzZXR9DQojIyMjIE1vZGVsDQpgYGB7cn0NCm1faGNfc2V4ZnJlcXBlbiA9IGJybShkaWFyeV9zZXhfYWN0aXZlX3NleF9zdW0gfg0KICAgICAgICAgICAgICAgICAgICAgICAgb2Zmc2V0KGxvZyhudW1iZXJfb2ZfZGF5cykpICsNCiAgICAgICAgICAgICAgICAgICAgICAgIGNvbnRyYWNlcHRpb25faG9ybW9uYWwsDQogICAgICAgICAgICAgICAgZGF0YSA9IGRhdGEsIGZhbWlseSA9IHBvaXNzb24oKSwNCiAgICAgICAgICAgICAgICBmaWxlID0gIm1faGNfc2V4ZnJlcXBlbiIpDQpgYGANCg0KIyMjIyBTdW1tYXJ5DQpgYGB7cn0NCnN1bW1hcnkobV9oY19zZXhmcmVxcGVuLCBpbnRlcnZhbHMgPSBULCBwcm9iID0gMC45MCkNCmBgYA0KDQojIyMjIENvbXBhcmlzb24gd2l0aCBST1BFDQpgYGB7cn0NCnBsb3QoZXF1aXZhbGVuY2VfdGVzdChtX2hjX3NleGZyZXFwZW4sIHJhbmdlID0gYygtMC4wNSwgMC4wNSkpLCBjaSA9IDAuOTAsDQogICAgICAgICAgICAgICAgICAgICAgcGFyYW1ldGVycyA9ICJjb250cmFjZXB0aW9uIikNCmVxdWl2YWxlbmNlX3Rlc3QobV9oY19zZXhmcmVxcGVuLCByYW5nZSA9IGMoLTAuMDUsIDAuMDUpLCBjaSA9IDAuOTAsDQogICAgICAgICAgICAgICAgICAgICAgcGFyYW1ldGVycyA9ICJjb250cmFjZXB0aW9uIikNCmBgYA0KDQojIyMjIFBsb3RzDQpgYGB7ciB3YXJuaW5nID0gRkFMU0V9DQpjb25kaXRpb25hbF9lZmZlY3RzKG1faGNfc2V4ZnJlcXBlbiwNCiAgICAgICAgICAgICAgICAgICAgZWZmZWN0cyA9ICJjb250cmFjZXB0aW9uX2hvcm1vbmFsIiwNCiAgICAgICAgICAgICAgICAgICAgY29uZGl0aW9ucyA9IGRhdGEuZnJhbWUobnVtYmVyX29mX2RheXMgPSAxKSkNCmBgYA0KDQojIyMjIEZvcmVzdCBQbG90IGZvciBFZmZlY3QgU2l6ZXMgey5hY3RpdmV9DQpgYGB7cn0NCm1faGNfc2V4ZnJlcXBlbiAlPiUNCiAgc3ByZWFkX2RyYXdzKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcykgJT4lDQogIHBpdm90X2xvbmdlcihjb2xzID0gYyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMpLA0KICAgICAgICAgICAgICAgbmFtZXNfdG8gPSAiY29uZGl0aW9uIiwNCiAgICAgICAgICAgICAgIHZhbHVlc190byA9ICJyX2NvbmRpdGlvbiIpICU+JQ0KICBtdXRhdGUoY29uZGl0aW9uX21lYW4gPSByX2NvbmRpdGlvbiwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnRyYWNlcHRpb24iLCBOQSksDQogICAgICAgICBjb25kaXRpb24gPSBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAiSG9ybW9uYWwgQ29udHJhY2VwdGlvbiIsIE5BKSkgJT4lDQogIGdncGxvdChhZXMoeSA9IGNvbmRpdGlvbiwNCiAgICAgICAgICAgICB4ID0gY29uZGl0aW9uX21lYW4sDQogICAgICAgICAgICAgZmlsbCA9IHN0YXQoYWJzKHgpIDwgMC4wNSkpKSArDQogIHN0YXRfaGFsZmV5ZSgpICsNCiAgZ2VvbV92bGluZSh4aW50ZXJjZXB0ID0gYygtMC4wNSwgMC4wNSksIGxpbmV0eXBlID0gImRvdHRlZCIpICsNCiAgYXBhdGhlbWUgKw0KICB0aGVtZShsZWdlbmQucG9zaXRpb24gPSAibm9uZSIpICsNCiAgc2NhbGVfZmlsbF9tYW51YWwodmFsdWVzID0gYygiZ3JheTgwIiwgInNreWJsdWUiKSkgKw0KICBsYWJzKHggPSAiRWZmZWN0IFNpemUgRXN0aW1hdGVzIiwgeSA9ICJQcmVkaWN0b3JzIikNCmBgYA0KDQojIyMgTWFzdHVyYmF0aW9uIEZyZXF1ZW5jeSB7LnRhYnNldH0NCiMjIyMgTW9kZWwNCmBgYHtyfQ0KbV9oY19tYXNmcmVxID0gYnJtKGRpYXJ5X21hc3R1cmJhdGlvbl9zdW0gfiBvZmZzZXQobG9nKG51bWJlcl9vZl9kYXlzKSkgKw0KICAgICAgICAgICAgICAgICAgICAgY29udHJhY2VwdGlvbl9ob3Jtb25hbCwNCiAgICAgICAgICAgICAgICBkYXRhID0gZGF0YSwgZmFtaWx5ID0gcG9pc3NvbigpLA0KICAgICAgICAgICAgICAgIGZpbGUgPSAibV9oY19tYXNmcmVxIikNCmBgYA0KDQojIyMjIFN1bW1hcnkNCmBgYHtyfQ0Kc3VtbWFyeShtX2hjX21hc2ZyZXEsIGludGVydmFscyA9IFQsIHByb2IgPSAwLjkwKQ0KYGBgDQoNCiMjIyMgQ29tcGFyaXNvbiB3aXRoIFJPUEUNCmBgYHtyfQ0KcGxvdChlcXVpdmFsZW5jZV90ZXN0KG1faGNfbWFzZnJlcSwgcmFuZ2UgPSBjKC0wLjA1LCAwLjA1KSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpKQ0KZXF1aXZhbGVuY2VfdGVzdChtX2hjX21hc2ZyZXEsIHJhbmdlID0gYygtMC4wNSwgMC4wNSksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKQ0KYGBgDQoNCiMjIyMgUGxvdHMNCmBgYHtyIHdhcm5pbmcgPSBGQUxTRX0NCmNvbmRpdGlvbmFsX2VmZmVjdHMobV9oY19tYXNmcmVxLA0KICAgICAgICAgICAgICAgICAgICBlZmZlY3RzID0gImNvbnRyYWNlcHRpb25faG9ybW9uYWwiLA0KICAgICAgICAgICAgICAgICAgICBjb25kaXRpb25zID0gZGF0YS5mcmFtZShudW1iZXJfb2ZfZGF5cyA9IDEpKQ0KYGBgDQoNCiMjIyMgRm9yZXN0IFBsb3QgZm9yIEVmZmVjdCBTaXplcyB7LmFjdGl2ZX0NCmBgYHtyfQ0KbV9oY19tYXNmcmVxICU+JQ0KICBzcHJlYWRfZHJhd3MoYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzKSAlPiUNCiAgcGl2b3RfbG9uZ2VyKGNvbHMgPSBjKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcyksDQogICAgICAgICAgICAgICBuYW1lc190byA9ICJjb25kaXRpb24iLA0KICAgICAgICAgICAgICAgdmFsdWVzX3RvID0gInJfY29uZGl0aW9uIikgJT4lDQogIG11dGF0ZShjb25kaXRpb25fbWVhbiA9IHJfY29uZGl0aW9uLA0KICAgICAgICAgZ3JvdXAgPSBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAiQ29udHJhY2VwdGlvbiIsIE5BKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAiYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJIb3Jtb25hbCBDb250cmFjZXB0aW9uIiwgTkEpKSAlPiUNCiAgZ2dwbG90KGFlcyh5ID0gY29uZGl0aW9uLA0KICAgICAgICAgICAgIHggPSBjb25kaXRpb25fbWVhbiwNCiAgICAgICAgICAgICBmaWxsID0gc3RhdChhYnMoeCkgPCAwLjA1KSkpICsNCiAgc3RhdF9oYWxmZXllKCkgKw0KICBnZW9tX3ZsaW5lKHhpbnRlcmNlcHQgPSBjKC0wLjA1LCAwLjA1KSwgbGluZXR5cGUgPSAiZG90dGVkIikgKw0KICBhcGF0aGVtZSArDQogIHRoZW1lKGxlZ2VuZC5wb3NpdGlvbiA9ICJub25lIikgKw0KICBzY2FsZV9maWxsX21hbnVhbCh2YWx1ZXMgPSBjKCJncmF5ODAiLCAic2t5Ymx1ZSIpKSArDQogIGxhYnMoeCA9ICJFZmZlY3QgU2l6ZSBFc3RpbWF0ZXMiLCB5ID0gIlByZWRpY3RvcnMiKQ0KYGBgDQoNCiMjIENvbnRyb2xsZWQgTW9kZWxzOiBFZmZlY3RzIG9mIEhvcm1vbmFsIENvbnRyYWNlcHRpdmVzIHsudGFic2V0fQ0KIyMjIEF0dHJhY3RpdmVuZXNzIG9mIFBhcnRuZXIgey50YWJzZXR9DQojIyMjIE1vZGVsDQpgYGB7cn0NCm1faGNfYXRycl9jb250cm9sbGVkID0gYnJtKGF0dHJhY3RpdmVuZXNzX3BhcnRuZXIgfiBjb250cmFjZXB0aW9uX2hvcm1vbmFsICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgYWdlICsgbmV0X2luY29tZSArIHJlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3IgKw0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgZWR1Y2F0aW9uX3llYXJzICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIGJmaV9leHRyYSArIGJmaV9uZXVybyArIGJmaV9hZ3JlZSArIGJmaV9jb25zYyArIGJmaV9vcGVuICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIHJlbGlnaW9zaXR5LA0KICAgICAgICAgICAgICAgIGRhdGEgPSBkYXRhLCBmYW1pbHkgPSBnYXVzc2lhbigpLA0KICAgICAgICAgICAgICAgIGZpbGUgPSAibV9oY19hdHJyX2NvbnRyb2xsZWQiKQ0KYGBgDQoNCiMjIyMgU3VtbWFyeQ0KYGBge3J9DQpzdW1tYXJ5KG1faGNfYXRycl9jb250cm9sbGVkLCBpbnRlcnZhbHMgPSBULCBwcm9iID0gMC45MCkNCmBgYA0KDQojIyMjIENvbXBhcmlzb24gd2l0aCBST1BFDQpgYGB7cn0NCnBsb3QoZXF1aXZhbGVuY2VfdGVzdChtX2hjX2F0cnJfY29udHJvbGxlZCwgcmFuZ2UgPSBjKC0wLjA3LCAwLjA3KSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpKQ0KZXF1aXZhbGVuY2VfdGVzdChtX2hjX2F0cnJfY29udHJvbGxlZCwgcmFuZ2UgPSBjKC0wLjA3LCAwLjA3KSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpDQpgYGANCg0KIyMjIyBQbG90cw0KYGBge3Igd2FybmluZyA9IEZBTFNFfQ0KY29uZGl0aW9uYWxfZWZmZWN0cyhtX2hjX2F0cnIsIGFzayA9IEZBTFNFKQ0KYGBgDQoNCiMjIyMgRm9yZXN0IFBsb3QgZm9yIEVmZmVjdCBTaXplcyB7LmFjdGl2ZX0NCmBgYHtyfQ0KbV9oY19hdHJyX2NvbnRyb2xsZWQgJT4lDQogIHNwcmVhZF9kcmF3cyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMsDQogICAgICAgICAgICAgICBiX2FnZSwNCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fNTAwXzEwMDAsIGJfbmV0X2luY29tZWV1cm9fMTAwMF8yMDAwLCANCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fMjAwMF8zMDAwLCBiX25ldF9pbmNvbWVldXJvX2d0XzMwMDAsIGJfbmV0X2luY29tZWRvbnRfdGVsbCwNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvMjhtb250aHMsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX21vcmV0aGFuNTJtb250aHMsDQogICAgICAgICAgICAgICBiX2VkdWNhdGlvbl95ZWFycywNCiAgICAgICAgICAgICAgIGJfYmZpX2V4dHJhLCBiX2JmaV9uZXVybywgYl9iZmlfYWdyZWUsIGJfYmZpX2NvbnNjLCBiX2JmaV9vcGVuLCANCiAgICAgICAgICAgICAgIGJfcmVsaWdpb3NpdHkpICU+JQ0KICBwaXZvdF9sb25nZXIoY29scyA9IGMoYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzLA0KICAgICAgICAgICAgICAgICAgICAgICAgYl9hZ2UsDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzUwMF8xMDAwLCBiX25ldF9pbmNvbWVldXJvXzEwMDBfMjAwMCwgDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzIwMDBfMzAwMCwgYl9uZXRfaW5jb21lZXVyb19ndF8zMDAwLCBiX25ldF9pbmNvbWVkb250X3RlbGwsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzI4bW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG81Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF9tb3JldGhhbjUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9lZHVjYXRpb25feWVhcnMsDQogICAgICAgICAgICAgICBiX2JmaV9leHRyYSwgYl9iZmlfbmV1cm8sIGJfYmZpX2FncmVlLCBiX2JmaV9jb25zYywgYl9iZmlfb3BlbiwgDQogICAgICAgICAgICAgICBiX3JlbGlnaW9zaXR5KSwNCiAgICAgICAgICAgICAgIG5hbWVzX3RvID0gImNvbmRpdGlvbiIsDQogICAgICAgICAgICAgICB2YWx1ZXNfdG8gPSAicl9jb25kaXRpb24iKSAlPiUNCiAgbXV0YXRlKGNvbmRpdGlvbl9tZWFuID0gcl9jb25kaXRpb24sDQogICAgICAgICBncm91cCA9IGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJSZWxhdGlvbnNoaXAgRHVyYXRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX25ldF9pbmNvbWUiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJJbmNvbWUiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIE5BKSksDQogICAgICAgICBncm91cCA9IGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAiYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJDb250cmFjZXB0aW9uIiwgZ3JvdXApLA0KICAgICAgICAgY29uZGl0aW9uID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLCBjb25kaXRpb24pLA0KICAgICAgICAgY29uZGl0aW9uID0gaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9hZ2UiLCAiQWdlIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVldXJvXzUwMF8xMDAwIiwgIjUwMC0xMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fMTAwMF8yMDAwIiwgIjEwMDAtMjAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVldXJvXzIwMDBfMzAwMCIsICIyMDAwLTMwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb19ndF8zMDAwIiwgIj4zMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWRvbnRfdGVsbCIsICJkbyBub3QgdGVsbCIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG8yOG1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICIxMy0yOCBtb250aHMiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvNTJtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAiMjktNTIgbW9udGhzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfbW9yZXRoYW41Mm1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICI+NTIgbW9udGhzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2VkdWNhdGlvbl95ZWFycyIsICJZZWFycyBvZiBFZHVjYXRpb24iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX2V4dHJhIiwgIkV4dHJhdmVyc2lvbiIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfbmV1cm8iLCAiTmV1cm90aWNpc20iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX2FncmVlIiwgIkFncmVlYWJsZW5lc3MiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX2NvbnNjIiwgIkNvbnNjaWVudGlvdXNuZXNzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9vcGVuIiwgIk9wZW5uZXNzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX3JlbGlnaW9zaXR5IiwgIlJlbGlnaW9zaXR5IiwNCiAgICAgICAgICAgICAgICAgICAgICAgY29uZGl0aW9uKSkpKSkpKSkpKSkpKSkpKSwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGlzLm5hKGdyb3VwKSwgY29uZGl0aW9uLCBncm91cCksDQogICAgICAgICBjb25kaXRpb24gPSBmYWN0b3IoY29uZGl0aW9uLCBsZXZlbHMgPSByZXYoYygiSG9ybW9uYWwgQ29udHJhY2VwdGlvbiIsICJBZ2UiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICI1MDAtMTAwMCBFdXJvIiwgIjEwMDAtMjAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiMjAwMC0zMDAwIEV1cm8iLCAiPjMwMDAgRXVybyIsICJkbyBub3QgdGVsbCIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIjEzLTI4IG1vbnRocyIsICIyOS01MiBtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICI+NTIgbW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiWWVhcnMgb2YgRWR1Y2F0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiRXh0cmF2ZXJzaW9uIiwgIk5ldXJvdGljaXNtIiwgIkFncmVlYWJsZW5lc3MiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiQ29uc2NpZW50aW91c25lc3MiLCJPcGVubmVzcyIsIlJlbGlnaW9zaXR5IikpKSwNCiAgICAgICAgIGdyb3VwID0gZmFjdG9yKGdyb3VwLCBsZXZlbHMgPSBjKCJDb250cmFjZXB0aW9uIiwgIkFnZSIsICJJbmNvbWUiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIlJlbGF0aW9uc2hpcCBEdXJhdGlvbiIsIlllYXJzIG9mIEVkdWNhdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiRXh0cmF2ZXJzaW9uIiwgIk5ldXJvdGljaXNtIiwgIkFncmVlYWJsZW5lc3MiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiQ29uc2NpZW50aW91c25lc3MiLCJPcGVubmVzcyIsIlJlbGlnaW9zaXR5IikpKSAlPiUNCiAgZ2dwbG90KGFlcyh5ID0gY29uZGl0aW9uLA0KICAgICAgICAgICAgIHggPSBjb25kaXRpb25fbWVhbiwNCiAgICAgICAgICAgICBmaWxsID0gc3RhdChhYnMoeCkgPCAwLjA3KSkpICsNCiAgc3RhdF9oYWxmZXllKCkgKw0KICBnZW9tX3ZsaW5lKHhpbnRlcmNlcHQgPSBjKC0wLjA3LCAwLjA3KSwgbGluZXR5cGUgPSAiZG90dGVkIikgKw0KICBhcGF0aGVtZSArDQogIHRoZW1lKGxlZ2VuZC5wb3NpdGlvbiA9ICJub25lIikgKw0KICBzY2FsZV9maWxsX21hbnVhbCh2YWx1ZXMgPSBjKCJncmF5ODAiLCAic2t5Ymx1ZSIpKSArDQogIGxhYnMoeCA9ICJFZmZlY3QgU2l6ZSBFc3RpbWF0ZXMiLCB5ID0gIlByZWRpY3RvcnMiKQ0KYGBgDQoNCiMjIyBSZWxhdGlvbnNoaXAgU2F0aXNmYWN0aW9uIHsudGFic2V0fQ0KIyMjIyBNb2RlbA0KYGBge3J9DQptX2hjX3JlbHNhdF9jb250cm9sbGVkID0gYnJtKHJlbGF0aW9uc2hpcF9zYXRpc2ZhY3Rpb24gfiBjb250cmFjZXB0aW9uX2hvcm1vbmFsICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgYWdlICsgbmV0X2luY29tZSArIHJlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3IgKw0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgZWR1Y2F0aW9uX3llYXJzICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIGJmaV9leHRyYSArIGJmaV9uZXVybyArIGJmaV9hZ3JlZSArIGJmaV9jb25zYyArIGJmaV9vcGVuICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIHJlbGlnaW9zaXR5LA0KICAgICAgICAgICAgICAgIGRhdGEgPSBkYXRhLCBmYW1pbHkgPSBnYXVzc2lhbigpLA0KICAgICAgICAgICAgICAgIGZpbGUgPSAibV9oY19yZWxzYXRfY29udHJvbGxlZCIpDQpgYGANCg0KIyMjIyBTdW1tYXJ5DQpgYGB7cn0NCnN1bW1hcnkobV9oY19yZWxzYXRfY29udHJvbGxlZCwgaW50ZXJ2YWxzID0gVCwgcHJvYiA9IDAuOTApDQpgYGANCg0KIyMjIyBDb21wYXJpc29uIHdpdGggUk9QRQ0KYGBge3J9DQpwbG90KGVxdWl2YWxlbmNlX3Rlc3QobV9oY19yZWxzYXRfY29udHJvbGxlZCwgcmFuZ2UgPSBjKC0wLjA0LCAwLjA0KSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpKQ0KDQplcXVpdmFsZW5jZV90ZXN0KG1faGNfcmVsc2F0X2NvbnRyb2xsZWQsIHJhbmdlID0gYygtMC4wNCwgMC4wNCksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKQ0KYGBgDQoNCiMjIyMgUGxvdHMNCmBgYHtyIHdhcm5pbmcgPSBGQUxTRX0NCmNvbmRpdGlvbmFsX2VmZmVjdHMobV9oY19yZWxzYXRfY29udHJvbGxlZCwgYXNrID0gRkFMU0UpDQpgYGANCg0KIyMjIyBGb3Jlc3QgUGxvdCBmb3IgRWZmZWN0IFNpemVzIHsuYWN0aXZlfQ0KYGBge3J9DQptX2hjX3JlbHNhdF9jb250cm9sbGVkICU+JQ0KICBzcHJlYWRfZHJhd3MoYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzLA0KICAgICAgICAgICAgICAgYl9hZ2UsDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzUwMF8xMDAwLCBiX25ldF9pbmNvbWVldXJvXzEwMDBfMjAwMCwgDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzIwMDBfMzAwMCwgYl9uZXRfaW5jb21lZXVyb19ndF8zMDAwLCBiX25ldF9pbmNvbWVkb250X3RlbGwsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzI4bW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG81Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF9tb3JldGhhbjUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9lZHVjYXRpb25feWVhcnMsDQogICAgICAgICAgICAgICBiX2JmaV9leHRyYSwgYl9iZmlfbmV1cm8sIGJfYmZpX2FncmVlLCBiX2JmaV9jb25zYywgYl9iZmlfb3BlbiwgDQogICAgICAgICAgICAgICBiX3JlbGlnaW9zaXR5KSAlPiUNCiAgcGl2b3RfbG9uZ2VyKGNvbHMgPSBjKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcywNCiAgICAgICAgICAgICAgICAgICAgICAgIGJfYWdlLA0KICAgICAgICAgICAgICAgYl9uZXRfaW5jb21lZXVyb181MDBfMTAwMCwgYl9uZXRfaW5jb21lZXVyb18xMDAwXzIwMDAsIA0KICAgICAgICAgICAgICAgYl9uZXRfaW5jb21lZXVyb18yMDAwXzMwMDAsIGJfbmV0X2luY29tZWV1cm9fZ3RfMzAwMCwgYl9uZXRfaW5jb21lZG9udF90ZWxsLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG8yOG1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvNTJtb250aHMsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfbW9yZXRoYW41Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfZWR1Y2F0aW9uX3llYXJzLA0KICAgICAgICAgICAgICAgYl9iZmlfZXh0cmEsIGJfYmZpX25ldXJvLCBiX2JmaV9hZ3JlZSwgYl9iZmlfY29uc2MsIGJfYmZpX29wZW4sIA0KICAgICAgICAgICAgICAgYl9yZWxpZ2lvc2l0eSksDQogICAgICAgICAgICAgICBuYW1lc190byA9ICJjb25kaXRpb24iLA0KICAgICAgICAgICAgICAgdmFsdWVzX3RvID0gInJfY29uZGl0aW9uIikgJT4lDQogIG11dGF0ZShjb25kaXRpb25fbWVhbiA9IHJfY29uZGl0aW9uLA0KICAgICAgICAgZ3JvdXAgPSBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvciIsDQogICAgICAgICAgICAgICAgICAgICAgICAiUmVsYXRpb25zaGlwIER1cmF0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAiYl9uZXRfaW5jb21lIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiSW5jb21lIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICBOQSkpLA0KICAgICAgICAgZ3JvdXAgPSBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAiQ29udHJhY2VwdGlvbiIsIGdyb3VwKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAiYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJIb3Jtb25hbCBDb250cmFjZXB0aW9uIiwgY29uZGl0aW9uKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGlmZWxzZShjb25kaXRpb24gPT0gImJfYWdlIiwgIkFnZSIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb181MDBfMTAwMCIsICI1MDAtMTAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVldXJvXzEwMDBfMjAwMCIsICIxMDAwLTIwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb18yMDAwXzMwMDAiLCAiMjAwMC0zMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fZ3RfMzAwMCIsICI+MzAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVkb250X3RlbGwiLCAiZG8gbm90IHRlbGwiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvMjhtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAiMTMtMjggbW9udGhzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzUybW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIjI5LTUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX21vcmV0aGFuNTJtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAiPjUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9lZHVjYXRpb25feWVhcnMiLCAiWWVhcnMgb2YgRWR1Y2F0aW9uIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9leHRyYSIsICJFeHRyYXZlcnNpb24iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX25ldXJvIiwgIk5ldXJvdGljaXNtIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9hZ3JlZSIsICJBZ3JlZWFibGVuZXNzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9jb25zYyIsICJDb25zY2llbnRpb3VzbmVzcyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfb3BlbiIsICJPcGVubmVzcyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxpZ2lvc2l0eSIsICJSZWxpZ2lvc2l0eSIsDQogICAgICAgICAgICAgICAgICAgICAgIGNvbmRpdGlvbikpKSkpKSkpKSkpKSkpKSksDQogICAgICAgICBncm91cCA9IGlmZWxzZShpcy5uYShncm91cCksIGNvbmRpdGlvbiwgZ3JvdXApLA0KICAgICAgICAgY29uZGl0aW9uID0gZmFjdG9yKGNvbmRpdGlvbiwgbGV2ZWxzID0gcmV2KGMoIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLCAiQWdlIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiNTAwLTEwMDAgRXVybyIsICIxMDAwLTIwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIjIwMDAtMzAwMCBFdXJvIiwgIj4zMDAwIEV1cm8iLCAiZG8gbm90IHRlbGwiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICIxMy0yOCBtb250aHMiLCAiMjktNTIgbW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiPjUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIlllYXJzIG9mIEVkdWNhdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkV4dHJhdmVyc2lvbiIsICJOZXVyb3RpY2lzbSIsICJBZ3JlZWFibGVuZXNzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnNjaWVudGlvdXNuZXNzIiwiT3Blbm5lc3MiLCJSZWxpZ2lvc2l0eSIpKSksDQogICAgICAgICBncm91cCA9IGZhY3Rvcihncm91cCwgbGV2ZWxzID0gYygiQ29udHJhY2VwdGlvbiIsICJBZ2UiLCAiSW5jb21lIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJSZWxhdGlvbnNoaXAgRHVyYXRpb24iLCJZZWFycyBvZiBFZHVjYXRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkV4dHJhdmVyc2lvbiIsICJOZXVyb3RpY2lzbSIsICJBZ3JlZWFibGVuZXNzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnNjaWVudGlvdXNuZXNzIiwiT3Blbm5lc3MiLCJSZWxpZ2lvc2l0eSIpKSkgJT4lDQogIGdncGxvdChhZXMoeSA9IGNvbmRpdGlvbiwNCiAgICAgICAgICAgICB4ID0gY29uZGl0aW9uX21lYW4sDQogICAgICAgICAgICAgZmlsbCA9IHN0YXQoYWJzKHgpIDwgMC4wNCkpKSArDQogIHN0YXRfaGFsZmV5ZSgpICsNCiAgZ2VvbV92bGluZSh4aW50ZXJjZXB0ID0gYygtMC4wNCwgMC4wNCksIGxpbmV0eXBlID0gImRvdHRlZCIpICsNCiAgYXBhdGhlbWUgKw0KICB0aGVtZShsZWdlbmQucG9zaXRpb24gPSAibm9uZSIpICsNCiAgc2NhbGVfZmlsbF9tYW51YWwodmFsdWVzID0gYygiZ3JheTgwIiwgInNreWJsdWUiKSkgKw0KICBsYWJzKHggPSAiRWZmZWN0IFNpemUgRXN0aW1hdGVzIiwgeSA9ICJQcmVkaWN0b3JzIikNCmBgYA0KDQoNCiMjIyBTZXh1YWwgU2F0aXNmYWN0aW9uIHsudGFic2V0fQ0KIyMjIyBNb2RlbA0KYGBge3J9DQptX2hjX3NleHNhdF9jb250cm9sbGVkID0gYnJtKHNhdGlzZmFjdGlvbl9zZXh1YWxfaW50ZXJjb3Vyc2UgfiBjb250cmFjZXB0aW9uX2hvcm1vbmFsICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgYWdlICsgbmV0X2luY29tZSArIHJlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3IgKw0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgZWR1Y2F0aW9uX3llYXJzICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIGJmaV9leHRyYSArIGJmaV9uZXVybyArIGJmaV9hZ3JlZSArIGJmaV9jb25zYyArIGJmaV9vcGVuICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIHJlbGlnaW9zaXR5LA0KICAgICAgICAgICAgICAgIGRhdGEgPSBkYXRhLCBmYW1pbHkgPSBnYXVzc2lhbigpLA0KICAgICAgICAgICAgICAgIGZpbGUgPSAibV9oY19zZXhzYXRfY29udHJvbGxlZCIpDQpgYGANCg0KIyMjIyBTdW1tYXJ5DQpgYGB7cn0NCnN1bW1hcnkobV9oY19zZXhzYXRfY29udHJvbGxlZCwgaW50ZXJ2YWxzID0gVCwgcHJvYiA9IDAuOTApDQpgYGANCg0KIyMjIyBDb21wYXJpc29uIHdpdGggUk9QRQ0KYGBge3J9DQpwbG90KGVxdWl2YWxlbmNlX3Rlc3QobV9oY19zZXhzYXRfY29udHJvbGxlZCwgcmFuZ2UgPSBjKC0wLjExLCAwLjExKSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpKQ0KZXF1aXZhbGVuY2VfdGVzdChtX2hjX3NleHNhdF9jb250cm9sbGVkLCByYW5nZSA9IGMoLTAuMTEsIDAuMTEpLCBjaSA9IDAuOTAsDQogICAgICAgICAgICAgICAgICAgICAgcGFyYW1ldGVycyA9ICJjb250cmFjZXB0aW9uIikNCmBgYA0KDQojIyMjIFBsb3RzDQpgYGB7ciB3YXJuaW5nID0gRkFMU0V9DQpjb25kaXRpb25hbF9lZmZlY3RzKG1faGNfc2V4c2F0X2NvbnRyb2xsZWQsIGFzayA9IEZBTFNFKQ0KYGBgDQoNCiMjIyMgRm9yZXN0IFBsb3QgZm9yIEVmZmVjdCBTaXplcyB7LmFjdGl2ZX0NCmBgYHtyfQ0KbV9oY19zZXhzYXRfY29udHJvbGxlZCAlPiUNCiAgc3ByZWFkX2RyYXdzKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcywNCiAgICAgICAgICAgICAgIGJfYWdlLA0KICAgICAgICAgICAgICAgYl9uZXRfaW5jb21lZXVyb181MDBfMTAwMCwgYl9uZXRfaW5jb21lZXVyb18xMDAwXzIwMDAsIA0KICAgICAgICAgICAgICAgYl9uZXRfaW5jb21lZXVyb18yMDAwXzMwMDAsIGJfbmV0X2luY29tZWV1cm9fZ3RfMzAwMCwgYl9uZXRfaW5jb21lZG9udF90ZWxsLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG8yOG1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvNTJtb250aHMsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfbW9yZXRoYW41Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfZWR1Y2F0aW9uX3llYXJzLA0KICAgICAgICAgICAgICAgYl9iZmlfZXh0cmEsIGJfYmZpX25ldXJvLCBiX2JmaV9hZ3JlZSwgYl9iZmlfY29uc2MsIGJfYmZpX29wZW4sIA0KICAgICAgICAgICAgICAgYl9yZWxpZ2lvc2l0eSkgJT4lDQogIHBpdm90X2xvbmdlcihjb2xzID0gYyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMsDQogICAgICAgICAgICAgICAgICAgICAgICBiX2FnZSwNCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fNTAwXzEwMDAsIGJfbmV0X2luY29tZWV1cm9fMTAwMF8yMDAwLCANCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fMjAwMF8zMDAwLCBiX25ldF9pbmNvbWVldXJvX2d0XzMwMDAsIGJfbmV0X2luY29tZWRvbnRfdGVsbCwNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvMjhtb250aHMsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX21vcmV0aGFuNTJtb250aHMsDQogICAgICAgICAgICAgICBiX2VkdWNhdGlvbl95ZWFycywNCiAgICAgICAgICAgICAgIGJfYmZpX2V4dHJhLCBiX2JmaV9uZXVybywgYl9iZmlfYWdyZWUsIGJfYmZpX2NvbnNjLCBiX2JmaV9vcGVuLCANCiAgICAgICAgICAgICAgIGJfcmVsaWdpb3NpdHkpLA0KICAgICAgICAgICAgICAgbmFtZXNfdG8gPSAiY29uZGl0aW9uIiwNCiAgICAgICAgICAgICAgIHZhbHVlc190byA9ICJyX2NvbmRpdGlvbiIpICU+JQ0KICBtdXRhdGUoY29uZGl0aW9uX21lYW4gPSByX2NvbmRpdGlvbiwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3IiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIlJlbGF0aW9uc2hpcCBEdXJhdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfbmV0X2luY29tZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkluY29tZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgTkEpKSwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnRyYWNlcHRpb24iLCBncm91cCksDQogICAgICAgICBjb25kaXRpb24gPSBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAiSG9ybW9uYWwgQ29udHJhY2VwdGlvbiIsIGNvbmRpdGlvbiksDQogICAgICAgICBjb25kaXRpb24gPSBpZmVsc2UoY29uZGl0aW9uID09ICJiX2FnZSIsICJBZ2UiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fNTAwXzEwMDAiLCAiNTAwLTEwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb18xMDAwXzIwMDAiLCAiMTAwMC0yMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fMjAwMF8zMDAwIiwgIjIwMDAtMzAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVldXJvX2d0XzMwMDAiLCAiPjMwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZG9udF90ZWxsIiwgImRvIG5vdCB0ZWxsIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzI4bW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIjEzLTI4IG1vbnRocyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG81Mm1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICIyOS01MiBtb250aHMiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF9tb3JldGhhbjUybW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIj41MiBtb250aHMiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfZWR1Y2F0aW9uX3llYXJzIiwgIlllYXJzIG9mIEVkdWNhdGlvbiIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfZXh0cmEiLCAiRXh0cmF2ZXJzaW9uIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9uZXVybyIsICJOZXVyb3RpY2lzbSIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfYWdyZWUiLCAiQWdyZWVhYmxlbmVzcyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfY29uc2MiLCAiQ29uc2NpZW50aW91c25lc3MiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX29wZW4iLCAiT3Blbm5lc3MiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsaWdpb3NpdHkiLCAiUmVsaWdpb3NpdHkiLA0KICAgICAgICAgICAgICAgICAgICAgICBjb25kaXRpb24pKSkpKSkpKSkpKSkpKSkpLA0KICAgICAgICAgZ3JvdXAgPSBpZmVsc2UoaXMubmEoZ3JvdXApLCBjb25kaXRpb24sIGdyb3VwKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGZhY3Rvcihjb25kaXRpb24sIGxldmVscyA9IHJldihjKCJIb3Jtb25hbCBDb250cmFjZXB0aW9uIiwgIkFnZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIjUwMC0xMDAwIEV1cm8iLCAiMTAwMC0yMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICIyMDAwLTMwMDAgRXVybyIsICI+MzAwMCBFdXJvIiwgImRvIG5vdCB0ZWxsIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiMTMtMjggbW9udGhzIiwgIjI5LTUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIj41MiBtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJZZWFycyBvZiBFZHVjYXRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJFeHRyYXZlcnNpb24iLCAiTmV1cm90aWNpc20iLCAiQWdyZWVhYmxlbmVzcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25zY2llbnRpb3VzbmVzcyIsIk9wZW5uZXNzIiwiUmVsaWdpb3NpdHkiKSkpLA0KICAgICAgICAgZ3JvdXAgPSBmYWN0b3IoZ3JvdXAsIGxldmVscyA9IGMoIkNvbnRyYWNlcHRpb24iLCAiQWdlIiwgIkluY29tZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiUmVsYXRpb25zaGlwIER1cmF0aW9uIiwiWWVhcnMgb2YgRWR1Y2F0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJFeHRyYXZlcnNpb24iLCAiTmV1cm90aWNpc20iLCAiQWdyZWVhYmxlbmVzcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25zY2llbnRpb3VzbmVzcyIsIk9wZW5uZXNzIiwiUmVsaWdpb3NpdHkiKSkpICU+JQ0KICBnZ3Bsb3QoYWVzKHkgPSBjb25kaXRpb24sDQogICAgICAgICAgICAgeCA9IGNvbmRpdGlvbl9tZWFuLA0KICAgICAgICAgICAgIGZpbGwgPSBzdGF0KGFicyh4KSA8IDAuMTEpKSkgKw0KICBzdGF0X2hhbGZleWUoKSArDQogIGdlb21fdmxpbmUoeGludGVyY2VwdCA9IGMoLTAuMTEsIDAuMTEpLCBsaW5ldHlwZSA9ICJkb3R0ZWQiKSArDQogIGFwYXRoZW1lICsNCiAgdGhlbWUobGVnZW5kLnBvc2l0aW9uID0gIm5vbmUiKSArDQogIHNjYWxlX2ZpbGxfbWFudWFsKHZhbHVlcyA9IGMoImdyYXk4MCIsICJza3libHVlIikpICsNCiAgbGFicyh4ID0gIkVmZmVjdCBTaXplIEVzdGltYXRlcyIsIHkgPSAiUHJlZGljdG9ycyIpDQpgYGANCg0KIyMjIExpYmlkbyB7LnRhYnNldH0NCg0KIyMjIyBNb2RlbA0KYGBge3J9DQptX2hjX2xpYmlkb19jb250cm9sbGVkID0gYnJtKGRpYXJ5X2xpYmlkb19tZWFuIH4gY29udHJhY2VwdGlvbl9ob3Jtb25hbCArDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgIGFnZSArIG5ldF9pbmNvbWUgKyByZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIGVkdWNhdGlvbl95ZWFycyArDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICBiZmlfZXh0cmEgKyBiZmlfbmV1cm8gKyBiZmlfYWdyZWUgKyBiZmlfY29uc2MgKyBiZmlfb3BlbiArDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICByZWxpZ2lvc2l0eSwNCiAgICAgICAgICAgICAgICBkYXRhID0gZGF0YSwgZmFtaWx5ID0gZ2F1c3NpYW4oKSwNCiAgICAgICAgICAgICAgICBmaWxlID0gIm1faGNfbGliaWRvX2NvbnRyb2xsZWQiKQ0KYGBgDQoNCiMjIyMgU3VtbWFyeQ0KYGBge3J9DQpzdW1tYXJ5KG1faGNfbGliaWRvX2NvbnRyb2xsZWQsIGludGVydmFscyA9IFQsIHByb2IgPSAwLjkwKQ0KYGBgDQoNCiMjIyMgQ29tcGFyaXNvbiB3aXRoIFJPUEUNCmBgYHtyfQ0KcGxvdChlcXVpdmFsZW5jZV90ZXN0KG1faGNfbGliaWRvX2NvbnRyb2xsZWQsIHJhbmdlID0gYygtMC4wNiwgMC4wNiksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKSkNCmVxdWl2YWxlbmNlX3Rlc3QobV9oY19saWJpZG9fY29udHJvbGxlZCwgcmFuZ2UgPSBjKC0wLjA2LCAwLjA2KSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpDQpgYGANCg0KIyMjIyBQbG90cw0KYGBge3Igd2FybmluZyA9IEZBTFNFfQ0KY29uZGl0aW9uYWxfZWZmZWN0cyhtX2hjX2xpYmlkb19jb250cm9sbGVkLCBhc2sgPSBGQUxTRSkNCmBgYA0KDQojIyMjIEZvcmVzdCBQbG90IGZvciBFZmZlY3QgU2l6ZXMgey5hY3RpdmV9DQpgYGB7cn0NCm1faGNfbGliaWRvX2NvbnRyb2xsZWQgJT4lDQogIHNwcmVhZF9kcmF3cyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMsDQogICAgICAgICAgICAgICBiX2FnZSwNCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fNTAwXzEwMDAsIGJfbmV0X2luY29tZWV1cm9fMTAwMF8yMDAwLCANCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fMjAwMF8zMDAwLCBiX25ldF9pbmNvbWVldXJvX2d0XzMwMDAsIGJfbmV0X2luY29tZWRvbnRfdGVsbCwNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvMjhtb250aHMsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX21vcmV0aGFuNTJtb250aHMsDQogICAgICAgICAgICAgICBiX2VkdWNhdGlvbl95ZWFycywNCiAgICAgICAgICAgICAgIGJfYmZpX2V4dHJhLCBiX2JmaV9uZXVybywgYl9iZmlfYWdyZWUsIGJfYmZpX2NvbnNjLCBiX2JmaV9vcGVuLCANCiAgICAgICAgICAgICAgIGJfcmVsaWdpb3NpdHkpICU+JQ0KICBwaXZvdF9sb25nZXIoY29scyA9IGMoYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzLA0KICAgICAgICAgICAgICAgICAgICAgICAgYl9hZ2UsDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzUwMF8xMDAwLCBiX25ldF9pbmNvbWVldXJvXzEwMDBfMjAwMCwgDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzIwMDBfMzAwMCwgYl9uZXRfaW5jb21lZXVyb19ndF8zMDAwLCBiX25ldF9pbmNvbWVkb250X3RlbGwsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzI4bW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG81Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF9tb3JldGhhbjUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9lZHVjYXRpb25feWVhcnMsDQogICAgICAgICAgICAgICBiX2JmaV9leHRyYSwgYl9iZmlfbmV1cm8sIGJfYmZpX2FncmVlLCBiX2JmaV9jb25zYywgYl9iZmlfb3BlbiwgDQogICAgICAgICAgICAgICBiX3JlbGlnaW9zaXR5KSwNCiAgICAgICAgICAgICAgIG5hbWVzX3RvID0gImNvbmRpdGlvbiIsDQogICAgICAgICAgICAgICB2YWx1ZXNfdG8gPSAicl9jb25kaXRpb24iKSAlPiUNCiAgbXV0YXRlKGNvbmRpdGlvbl9tZWFuID0gcl9jb25kaXRpb24sDQogICAgICAgICBncm91cCA9IGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJSZWxhdGlvbnNoaXAgRHVyYXRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX25ldF9pbmNvbWUiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJJbmNvbWUiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIE5BKSksDQogICAgICAgICBncm91cCA9IGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAiYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJDb250cmFjZXB0aW9uIiwgZ3JvdXApLA0KICAgICAgICAgY29uZGl0aW9uID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLCBjb25kaXRpb24pLA0KICAgICAgICAgY29uZGl0aW9uID0gaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9hZ2UiLCAiQWdlIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVldXJvXzUwMF8xMDAwIiwgIjUwMC0xMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fMTAwMF8yMDAwIiwgIjEwMDAtMjAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVldXJvXzIwMDBfMzAwMCIsICIyMDAwLTMwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb19ndF8zMDAwIiwgIj4zMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWRvbnRfdGVsbCIsICJkbyBub3QgdGVsbCIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG8yOG1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICIxMy0yOCBtb250aHMiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvNTJtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAiMjktNTIgbW9udGhzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfbW9yZXRoYW41Mm1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICI+NTIgbW9udGhzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2VkdWNhdGlvbl95ZWFycyIsICJZZWFycyBvZiBFZHVjYXRpb24iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX2V4dHJhIiwgIkV4dHJhdmVyc2lvbiIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfbmV1cm8iLCAiTmV1cm90aWNpc20iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX2FncmVlIiwgIkFncmVlYWJsZW5lc3MiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX2NvbnNjIiwgIkNvbnNjaWVudGlvdXNuZXNzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9vcGVuIiwgIk9wZW5uZXNzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX3JlbGlnaW9zaXR5IiwgIlJlbGlnaW9zaXR5IiwNCiAgICAgICAgICAgICAgICAgICAgICAgY29uZGl0aW9uKSkpKSkpKSkpKSkpKSkpKSwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGlzLm5hKGdyb3VwKSwgY29uZGl0aW9uLCBncm91cCksDQogICAgICAgICBjb25kaXRpb24gPSBmYWN0b3IoY29uZGl0aW9uLCBsZXZlbHMgPSByZXYoYygiSG9ybW9uYWwgQ29udHJhY2VwdGlvbiIsICJBZ2UiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICI1MDAtMTAwMCBFdXJvIiwgIjEwMDAtMjAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiMjAwMC0zMDAwIEV1cm8iLCAiPjMwMDAgRXVybyIsICJkbyBub3QgdGVsbCIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIjEzLTI4IG1vbnRocyIsICIyOS01MiBtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICI+NTIgbW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiWWVhcnMgb2YgRWR1Y2F0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiRXh0cmF2ZXJzaW9uIiwgIk5ldXJvdGljaXNtIiwgIkFncmVlYWJsZW5lc3MiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiQ29uc2NpZW50aW91c25lc3MiLCJPcGVubmVzcyIsIlJlbGlnaW9zaXR5IikpKSwNCiAgICAgICAgIGdyb3VwID0gZmFjdG9yKGdyb3VwLCBsZXZlbHMgPSBjKCJDb250cmFjZXB0aW9uIiwgIkFnZSIsICJJbmNvbWUiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIlJlbGF0aW9uc2hpcCBEdXJhdGlvbiIsIlllYXJzIG9mIEVkdWNhdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiRXh0cmF2ZXJzaW9uIiwgIk5ldXJvdGljaXNtIiwgIkFncmVlYWJsZW5lc3MiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiQ29uc2NpZW50aW91c25lc3MiLCJPcGVubmVzcyIsIlJlbGlnaW9zaXR5IikpKSAlPiUNCiAgZ2dwbG90KGFlcyh5ID0gY29uZGl0aW9uLA0KICAgICAgICAgICAgIHggPSBjb25kaXRpb25fbWVhbiwNCiAgICAgICAgICAgICBmaWxsID0gc3RhdChhYnMoeCkgPCAwLjA2KSkpICsNCiAgc3RhdF9oYWxmZXllKCkgKw0KICBnZW9tX3ZsaW5lKHhpbnRlcmNlcHQgPSBjKC0wLjA2LCAwLjA2KSwgbGluZXR5cGUgPSAiZG90dGVkIikgKw0KICBhcGF0aGVtZSArDQogIHRoZW1lKGxlZ2VuZC5wb3NpdGlvbiA9ICJub25lIikgKw0KICBzY2FsZV9maWxsX21hbnVhbCh2YWx1ZXMgPSBjKCJncmF5ODAiLCAic2t5Ymx1ZSIpKSArDQogIGxhYnMoeCA9ICJFZmZlY3QgU2l6ZSBFc3RpbWF0ZXMiLCB5ID0gIlByZWRpY3RvcnMiKQ0KYGBgDQoNCiMjIyBTZXh1YWwgRnJlcXVlbmN5IChQZW5ldHJhdGl2ZSBJbnRlcmNvdXJzZSkgey50YWJzZXR9DQojIyMjIE1vZGVsDQpgYGB7cn0NCm1faGNfc2V4ZnJlcXBlbl9jb250cm9sbGVkID0gYnJtKGRpYXJ5X3NleF9hY3RpdmVfc2V4X3N1bSB+DQogICAgICAgICAgICAgICAgICAgICAgICBvZmZzZXQobG9nKG51bWJlcl9vZl9kYXlzKSkgKw0KICAgICAgICAgICAgICAgICAgICAgICAgY29udHJhY2VwdGlvbl9ob3Jtb25hbCArDQogICAgICAgICAgICAgICAgICAgICAgICBhZ2UgKyBuZXRfaW5jb21lICsgcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvciArDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICBlZHVjYXRpb25feWVhcnMgKw0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgYmZpX2V4dHJhICsgYmZpX25ldXJvICsgYmZpX2FncmVlICsgYmZpX2NvbnNjICsgYmZpX29wZW4gKw0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgcmVsaWdpb3NpdHksDQogICAgICAgICAgICAgICAgZGF0YSA9IGRhdGEsIGZhbWlseSA9IHBvaXNzb24oKSwNCiAgICAgICAgICAgICAgICBmaWxlID0gIm1faGNfc2V4ZnJlcXBlbl9jb250cm9sbGVkIikNCmBgYA0KDQojIyMjIFN1bW1hcnkNCmBgYHtyfQ0Kc3VtbWFyeShtX2hjX3NleGZyZXFwZW5fY29udHJvbGxlZCwgaW50ZXJ2YWxzID0gVCwgcHJvYiA9IDAuOTApDQpgYGANCg0KIyMjIyBDb21wYXJpc29uIHdpdGggUk9QRQ0KYGBge3J9DQpwbG90KGVxdWl2YWxlbmNlX3Rlc3QobV9oY19zZXhmcmVxcGVuX2NvbnRyb2xsZWQsIHJhbmdlID0gYygtMC4wNSwgMC4wNSksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKSkNCmVxdWl2YWxlbmNlX3Rlc3QobV9oY19zZXhmcmVxcGVuX2NvbnRyb2xsZWQsIHJhbmdlID0gYygtMC4wNSwgMC4wNSksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKQ0KYGBgDQoNCiMjIyMgUGxvdHMNCmBgYHtyIHdhcm5pbmcgPSBGQUxTRX0NCmNvbmRpdGlvbmFsX2VmZmVjdHMobV9oY19zZXhmcmVxcGVuX2NvbnRyb2xsZWQsDQogICAgICAgICAgICAgICAgICAgIGVmZmVjdHMgPSAiY29udHJhY2VwdGlvbl9ob3Jtb25hbCIsDQogICAgICAgICAgICAgICAgICAgIGNvbmRpdGlvbnMgPSBkYXRhLmZyYW1lKG51bWJlcl9vZl9kYXlzID0gMSkpDQpgYGANCg0KIyMjIyBGb3Jlc3QgUGxvdCBmb3IgRWZmZWN0IFNpemVzIHsuYWN0aXZlfQ0KYGBge3J9DQptX2hjX3NleGZyZXFwZW5fY29udHJvbGxlZCAlPiUNCiAgc3ByZWFkX2RyYXdzKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcywNCiAgICAgICAgICAgICAgIGJfYWdlLA0KICAgICAgICAgICAgICAgYl9uZXRfaW5jb21lZXVyb181MDBfMTAwMCwgYl9uZXRfaW5jb21lZXVyb18xMDAwXzIwMDAsIA0KICAgICAgICAgICAgICAgYl9uZXRfaW5jb21lZXVyb18yMDAwXzMwMDAsIGJfbmV0X2luY29tZWV1cm9fZ3RfMzAwMCwgYl9uZXRfaW5jb21lZG9udF90ZWxsLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG8yOG1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvNTJtb250aHMsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfbW9yZXRoYW41Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfZWR1Y2F0aW9uX3llYXJzLA0KICAgICAgICAgICAgICAgYl9iZmlfZXh0cmEsIGJfYmZpX25ldXJvLCBiX2JmaV9hZ3JlZSwgYl9iZmlfY29uc2MsIGJfYmZpX29wZW4sIA0KICAgICAgICAgICAgICAgYl9yZWxpZ2lvc2l0eSkgJT4lDQogIHBpdm90X2xvbmdlcihjb2xzID0gYyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMsDQogICAgICAgICAgICAgICAgICAgICAgICBiX2FnZSwNCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fNTAwXzEwMDAsIGJfbmV0X2luY29tZWV1cm9fMTAwMF8yMDAwLCANCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fMjAwMF8zMDAwLCBiX25ldF9pbmNvbWVldXJvX2d0XzMwMDAsIGJfbmV0X2luY29tZWRvbnRfdGVsbCwNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvMjhtb250aHMsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX21vcmV0aGFuNTJtb250aHMsDQogICAgICAgICAgICAgICBiX2VkdWNhdGlvbl95ZWFycywNCiAgICAgICAgICAgICAgIGJfYmZpX2V4dHJhLCBiX2JmaV9uZXVybywgYl9iZmlfYWdyZWUsIGJfYmZpX2NvbnNjLCBiX2JmaV9vcGVuLCANCiAgICAgICAgICAgICAgIGJfcmVsaWdpb3NpdHkpLA0KICAgICAgICAgICAgICAgbmFtZXNfdG8gPSAiY29uZGl0aW9uIiwNCiAgICAgICAgICAgICAgIHZhbHVlc190byA9ICJyX2NvbmRpdGlvbiIpICU+JQ0KICBtdXRhdGUoY29uZGl0aW9uX21lYW4gPSByX2NvbmRpdGlvbiwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3IiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIlJlbGF0aW9uc2hpcCBEdXJhdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfbmV0X2luY29tZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkluY29tZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgTkEpKSwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnRyYWNlcHRpb24iLCBncm91cCksDQogICAgICAgICBjb25kaXRpb24gPSBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAiSG9ybW9uYWwgQ29udHJhY2VwdGlvbiIsIGNvbmRpdGlvbiksDQogICAgICAgICBjb25kaXRpb24gPSBpZmVsc2UoY29uZGl0aW9uID09ICJiX2FnZSIsICJBZ2UiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fNTAwXzEwMDAiLCAiNTAwLTEwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb18xMDAwXzIwMDAiLCAiMTAwMC0yMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fMjAwMF8zMDAwIiwgIjIwMDAtMzAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVldXJvX2d0XzMwMDAiLCAiPjMwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZG9udF90ZWxsIiwgImRvIG5vdCB0ZWxsIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzI4bW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIjEzLTI4IG1vbnRocyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG81Mm1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICIyOS01MiBtb250aHMiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF9tb3JldGhhbjUybW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIj41MiBtb250aHMiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfZWR1Y2F0aW9uX3llYXJzIiwgIlllYXJzIG9mIEVkdWNhdGlvbiIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfZXh0cmEiLCAiRXh0cmF2ZXJzaW9uIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9uZXVybyIsICJOZXVyb3RpY2lzbSIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfYWdyZWUiLCAiQWdyZWVhYmxlbmVzcyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfY29uc2MiLCAiQ29uc2NpZW50aW91c25lc3MiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX29wZW4iLCAiT3Blbm5lc3MiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsaWdpb3NpdHkiLCAiUmVsaWdpb3NpdHkiLA0KICAgICAgICAgICAgICAgICAgICAgICBjb25kaXRpb24pKSkpKSkpKSkpKSkpKSkpLA0KICAgICAgICAgZ3JvdXAgPSBpZmVsc2UoaXMubmEoZ3JvdXApLCBjb25kaXRpb24sIGdyb3VwKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGZhY3Rvcihjb25kaXRpb24sIGxldmVscyA9IHJldihjKCJIb3Jtb25hbCBDb250cmFjZXB0aW9uIiwgIkFnZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIjUwMC0xMDAwIEV1cm8iLCAiMTAwMC0yMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICIyMDAwLTMwMDAgRXVybyIsICI+MzAwMCBFdXJvIiwgImRvIG5vdCB0ZWxsIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiMTMtMjggbW9udGhzIiwgIjI5LTUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIj41MiBtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJZZWFycyBvZiBFZHVjYXRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJFeHRyYXZlcnNpb24iLCAiTmV1cm90aWNpc20iLCAiQWdyZWVhYmxlbmVzcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25zY2llbnRpb3VzbmVzcyIsIk9wZW5uZXNzIiwiUmVsaWdpb3NpdHkiKSkpLA0KICAgICAgICAgZ3JvdXAgPSBmYWN0b3IoZ3JvdXAsIGxldmVscyA9IGMoIkNvbnRyYWNlcHRpb24iLCAiQWdlIiwgIkluY29tZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiUmVsYXRpb25zaGlwIER1cmF0aW9uIiwiWWVhcnMgb2YgRWR1Y2F0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJFeHRyYXZlcnNpb24iLCAiTmV1cm90aWNpc20iLCAiQWdyZWVhYmxlbmVzcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25zY2llbnRpb3VzbmVzcyIsIk9wZW5uZXNzIiwiUmVsaWdpb3NpdHkiKSkpICU+JQ0KICBnZ3Bsb3QoYWVzKHkgPSBjb25kaXRpb24sDQogICAgICAgICAgICAgeCA9IGNvbmRpdGlvbl9tZWFuLA0KICAgICAgICAgICAgIGZpbGwgPSBzdGF0KGFicyh4KSA8IDAuMDUpKSkgKw0KICBzdGF0X2hhbGZleWUoKSArDQogIGdlb21fdmxpbmUoeGludGVyY2VwdCA9IGMoLTAuMDUsIDAuMDUpLCBsaW5ldHlwZSA9ICJkb3R0ZWQiKSArDQogIGFwYXRoZW1lICsNCiAgdGhlbWUobGVnZW5kLnBvc2l0aW9uID0gIm5vbmUiKSArDQogIHNjYWxlX2ZpbGxfbWFudWFsKHZhbHVlcyA9IGMoImdyYXk4MCIsICJza3libHVlIikpICsNCiAgbGFicyh4ID0gIkVmZmVjdCBTaXplIEVzdGltYXRlcyIsIHkgPSAiUHJlZGljdG9ycyIpDQpgYGANCg0KIyMjIE1hc3R1cmFiYXRpb24gRnJlcXVlbmN5IHsudGFic2V0fQ0KIyMjIyBNb2RlbA0KYGBge3J9DQptX2hjX21hc2ZyZXFfY29udHJvbGxlZCA9IGJybShkaWFyeV9tYXN0dXJiYXRpb25fc3VtIH4NCiAgICAgICAgICAgICAgICAgICAgICAgIG9mZnNldChsb2cobnVtYmVyX29mX2RheXMpKSArDQogICAgICAgICAgICAgICAgICAgICAgICBjb250cmFjZXB0aW9uX2hvcm1vbmFsICsNCiAgICAgICAgICAgICAgICAgICAgICAgIGFnZSArIG5ldF9pbmNvbWUgKyByZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIGVkdWNhdGlvbl95ZWFycyArDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICBiZmlfZXh0cmEgKyBiZmlfbmV1cm8gKyBiZmlfYWdyZWUgKyBiZmlfY29uc2MgKyBiZmlfb3BlbiArDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICByZWxpZ2lvc2l0eSwNCiAgICAgICAgICAgICAgICBkYXRhID0gZGF0YSwgZmFtaWx5ID0gcG9pc3NvbigpLA0KICAgICAgICAgICAgICAgIGZpbGUgPSAibV9oY19tYXNmcmVxX2NvbnRyb2xsZWQiKQ0KYGBgDQoNCiMjIyMgU3VtbWFyeQ0KYGBge3J9DQpzdW1tYXJ5KG1faGNfbWFzZnJlcV9jb250cm9sbGVkLCBpbnRlcnZhbHMgPSBULCBwcm9iID0gMC45MCkNCmBgYA0KDQojIyMjIENvbXBhcmlzb24gd2l0aCBST1BFDQpgYGB7cn0NCnBsb3QoZXF1aXZhbGVuY2VfdGVzdChtX2hjX21hc2ZyZXFfY29udHJvbGxlZCwgcmFuZ2UgPSBjKC0wLjA1LCAwLjA1KSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpKQ0KZXF1aXZhbGVuY2VfdGVzdChtX2hjX21hc2ZyZXFfY29udHJvbGxlZCwgcmFuZ2UgPSBjKC0wLjA1LCAwLjA1KSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpDQpgYGANCg0KIyMjIyBQbG90cw0KYGBge3Igd2FybmluZyA9IEZBTFNFfQ0KY29uZGl0aW9uYWxfZWZmZWN0cyhtX2hjX21hc2ZyZXFfY29udHJvbGxlZCwNCiAgICAgICAgICAgICAgICAgICAgZWZmZWN0cyA9ICJjb250cmFjZXB0aW9uX2hvcm1vbmFsIiwNCiAgICAgICAgICAgICAgICAgICAgY29uZGl0aW9ucyA9IGRhdGEuZnJhbWUobnVtYmVyX29mX2RheXMgPSAxKSkNCmBgYA0KDQojIyMjIEZvcmVzdCBQbG90IGZvciBFZmZlY3QgU2l6ZXMgey5hY3RpdmV9DQpgYGB7cn0NCm1faGNfbWFzZnJlcV9jb250cm9sbGVkICU+JQ0KICBzcHJlYWRfZHJhd3MoYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzLA0KICAgICAgICAgICAgICAgYl9hZ2UsDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzUwMF8xMDAwLCBiX25ldF9pbmNvbWVldXJvXzEwMDBfMjAwMCwgDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzIwMDBfMzAwMCwgYl9uZXRfaW5jb21lZXVyb19ndF8zMDAwLCBiX25ldF9pbmNvbWVkb250X3RlbGwsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzI4bW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG81Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF9tb3JldGhhbjUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9lZHVjYXRpb25feWVhcnMsDQogICAgICAgICAgICAgICBiX2JmaV9leHRyYSwgYl9iZmlfbmV1cm8sIGJfYmZpX2FncmVlLCBiX2JmaV9jb25zYywgYl9iZmlfb3BlbiwgDQogICAgICAgICAgICAgICBiX3JlbGlnaW9zaXR5KSAlPiUNCiAgcGl2b3RfbG9uZ2VyKGNvbHMgPSBjKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcywNCiAgICAgICAgICAgICAgICAgICAgICAgIGJfYWdlLA0KICAgICAgICAgICAgICAgYl9uZXRfaW5jb21lZXVyb181MDBfMTAwMCwgYl9uZXRfaW5jb21lZXVyb18xMDAwXzIwMDAsIA0KICAgICAgICAgICAgICAgYl9uZXRfaW5jb21lZXVyb18yMDAwXzMwMDAsIGJfbmV0X2luY29tZWV1cm9fZ3RfMzAwMCwgYl9uZXRfaW5jb21lZG9udF90ZWxsLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG8yOG1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvNTJtb250aHMsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfbW9yZXRoYW41Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfZWR1Y2F0aW9uX3llYXJzLA0KICAgICAgICAgICAgICAgYl9iZmlfZXh0cmEsIGJfYmZpX25ldXJvLCBiX2JmaV9hZ3JlZSwgYl9iZmlfY29uc2MsIGJfYmZpX29wZW4sIA0KICAgICAgICAgICAgICAgYl9yZWxpZ2lvc2l0eSksDQogICAgICAgICAgICAgICBuYW1lc190byA9ICJjb25kaXRpb24iLA0KICAgICAgICAgICAgICAgdmFsdWVzX3RvID0gInJfY29uZGl0aW9uIikgJT4lDQogIG11dGF0ZShjb25kaXRpb25fbWVhbiA9IHJfY29uZGl0aW9uLA0KICAgICAgICAgZ3JvdXAgPSBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvciIsDQogICAgICAgICAgICAgICAgICAgICAgICAiUmVsYXRpb25zaGlwIER1cmF0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAiYl9uZXRfaW5jb21lIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiSW5jb21lIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICBOQSkpLA0KICAgICAgICAgZ3JvdXAgPSBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAiQ29udHJhY2VwdGlvbiIsIGdyb3VwKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAiYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJIb3Jtb25hbCBDb250cmFjZXB0aW9uIiwgY29uZGl0aW9uKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGlmZWxzZShjb25kaXRpb24gPT0gImJfYWdlIiwgIkFnZSIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb181MDBfMTAwMCIsICI1MDAtMTAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVldXJvXzEwMDBfMjAwMCIsICIxMDAwLTIwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb18yMDAwXzMwMDAiLCAiMjAwMC0zMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fZ3RfMzAwMCIsICI+MzAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVkb250X3RlbGwiLCAiZG8gbm90IHRlbGwiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvMjhtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAiMTMtMjggbW9udGhzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzUybW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIjI5LTUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX21vcmV0aGFuNTJtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAiPjUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9lZHVjYXRpb25feWVhcnMiLCAiWWVhcnMgb2YgRWR1Y2F0aW9uIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9leHRyYSIsICJFeHRyYXZlcnNpb24iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX25ldXJvIiwgIk5ldXJvdGljaXNtIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9hZ3JlZSIsICJBZ3JlZWFibGVuZXNzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9jb25zYyIsICJDb25zY2llbnRpb3VzbmVzcyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfb3BlbiIsICJPcGVubmVzcyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxpZ2lvc2l0eSIsICJSZWxpZ2lvc2l0eSIsDQogICAgICAgICAgICAgICAgICAgICAgIGNvbmRpdGlvbikpKSkpKSkpKSkpKSkpKSksDQogICAgICAgICBncm91cCA9IGlmZWxzZShpcy5uYShncm91cCksIGNvbmRpdGlvbiwgZ3JvdXApLA0KICAgICAgICAgY29uZGl0aW9uID0gZmFjdG9yKGNvbmRpdGlvbiwgbGV2ZWxzID0gcmV2KGMoIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLCAiQWdlIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiNTAwLTEwMDAgRXVybyIsICIxMDAwLTIwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIjIwMDAtMzAwMCBFdXJvIiwgIj4zMDAwIEV1cm8iLCAiZG8gbm90IHRlbGwiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICIxMy0yOCBtb250aHMiLCAiMjktNTIgbW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiPjUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIlllYXJzIG9mIEVkdWNhdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkV4dHJhdmVyc2lvbiIsICJOZXVyb3RpY2lzbSIsICJBZ3JlZWFibGVuZXNzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnNjaWVudGlvdXNuZXNzIiwiT3Blbm5lc3MiLCJSZWxpZ2lvc2l0eSIpKSksDQogICAgICAgICBncm91cCA9IGZhY3Rvcihncm91cCwgbGV2ZWxzID0gYygiQ29udHJhY2VwdGlvbiIsICJBZ2UiLCAiSW5jb21lIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJSZWxhdGlvbnNoaXAgRHVyYXRpb24iLCJZZWFycyBvZiBFZHVjYXRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkV4dHJhdmVyc2lvbiIsICJOZXVyb3RpY2lzbSIsICJBZ3JlZWFibGVuZXNzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnNjaWVudGlvdXNuZXNzIiwiT3Blbm5lc3MiLCJSZWxpZ2lvc2l0eSIpKSkgJT4lDQogIGdncGxvdChhZXMoeSA9IGNvbmRpdGlvbiwNCiAgICAgICAgICAgICB4ID0gY29uZGl0aW9uX21lYW4sDQogICAgICAgICAgICAgZmlsbCA9IHN0YXQoYWJzKHgpIDwgMC4wNSkpKSArDQogIHN0YXRfaGFsZmV5ZSgpICsNCiAgZ2VvbV92bGluZSh4aW50ZXJjZXB0ID0gYygtMC4wNSwgMC4wNSksIGxpbmV0eXBlID0gImRvdHRlZCIpICsNCiAgYXBhdGhlbWUgKw0KICB0aGVtZShsZWdlbmQucG9zaXRpb24gPSAibm9uZSIpICsNCiAgc2NhbGVfZmlsbF9tYW51YWwodmFsdWVzID0gYygiZ3JheTgwIiwgInNreWJsdWUiKSkgKw0KICBsYWJzKHggPSAiRWZmZWN0IFNpemUgRXN0aW1hdGVzIiwgeSA9ICJQcmVkaWN0b3JzIikNCmBgYA0KDQojIyBFZmZlY3RzIG9mIChJbilDb25ncnVlbmN0IEhDIFVzZSB7LnRhYnNldH0NCiMjIyBBdHRyYWN0aXZlbmVzcyBvZiBQYXJ0bmVyICB7LnRhYnNldH0NCiMjIyMgTW9kZWwNCmBgYHtyfQ0KbV9jb25ncnVlbmN5X2F0cnIgPSBicm0oYXR0cmFjdGl2ZW5lc3NfcGFydG5lciB+DQogICAgICAgICAgICAgICAgICAgICAgICAgIGNvbnRyYWNlcHRpb25faG9ybW9uYWwgKiBjb25ncnVlbnRfY29udHJhY2VwdGlvbiwNCiAgICAgICAgICAgICAgICBkYXRhID0gZGF0YSwgZmFtaWx5ID0gZ2F1c3NpYW4oKSwNCiAgICAgICAgICAgICAgICBmaWxlID0gIm1fY29uZ3J1ZW5jeV9hdHJyIikNCmBgYA0KDQojIyMjIFN1bW1hcnkNCmBgYHtyfQ0Kc3VtbWFyeShtX2NvbmdydWVuY3lfYXRyciwgaW50ZXJ2YWxzID0gVCwgcHJvYiA9IDAuOTApDQpgYGANCg0KIyMjIyBDb21wYXJpc29uIHdpdGggUk9QRQ0KYGBge3J9DQpwbG90KGVxdWl2YWxlbmNlX3Rlc3QobV9jb25ncnVlbmN5X2F0cnIsIHJhbmdlID0gYygtMC4wNywgMC4wNyksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKSkNCmVxdWl2YWxlbmNlX3Rlc3QobV9jb25ncnVlbmN5X2F0cnIsIHJhbmdlID0gYygtMC4wNywgMC4wNyksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKQ0KYGBgDQoNCiMjIyMgUGxvdHMNCmBgYHtyIHdhcm5pbmcgPSBGQUxTRX0NCmNvbmRpdGlvbmFsX2VmZmVjdHMobV9jb25ncnVlbmN5X2F0cnIsIGFzayA9IEZBTFNFKQ0KYGBgDQoNCiMjIyMgRm9yZXN0IFBsb3QgZm9yIEVmZmVjdCBTaXplcyB7LmFjdGl2ZX0NCmBgYHtyfQ0KbV9jb25ncnVlbmN5X2F0cnIgJT4lDQogIHNwcmVhZF9kcmF3cyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMsIGJfY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xLA0KICAgICAgICAgICAgICAgYGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllczpjb25ncnVlbnRfY29udHJhY2VwdGlvbjFgKSAlPiUNCiAgcGl2b3RfbG9uZ2VyKGNvbHMgPSBjKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcywgYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEsDQogICAgICAgICAgICAgICBgYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzOmNvbmdydWVudF9jb250cmFjZXB0aW9uMWApLA0KICAgICAgICAgICAgICAgbmFtZXNfdG8gPSAiY29uZGl0aW9uIiwNCiAgICAgICAgICAgICAgIHZhbHVlc190byA9ICJyX2NvbmRpdGlvbiIpICU+JQ0KICBtdXRhdGUoY29uZGl0aW9uX21lYW4gPSByX2NvbmRpdGlvbiwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnRyYWNlcHRpb24iLCBOQSksDQogICAgICAgICBjb25kaXRpb24gPSBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXM6Y29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAiSW50ZXJhY3Rpb24gSG9ybW9uYWwgQ29udHJhY3BldGlvbiBhbmQgQ29uZ3J1ZW50IENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgIGNvbmRpdGlvbikpKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGZhY3Rvcihjb25kaXRpb24sIGxldmVscyA9IHJldihjKCJIb3Jtb25hbCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbmdydWVudCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkludGVyYWN0aW9uIEhvcm1vbmFsIENvbnRyYWNwZXRpb24gYW5kIENvbmdydWVudCBDb250cmFjZXB0aW9uIikpKSkgJT4lDQogIGdncGxvdChhZXMoeSA9IGNvbmRpdGlvbiwNCiAgICAgICAgICAgICB4ID0gY29uZGl0aW9uX21lYW4sDQogICAgICAgICAgICAgZmlsbCA9IHN0YXQoYWJzKHgpIDwgMC4wNykpKSArDQogIHN0YXRfaGFsZmV5ZSgpICsNCiAgZ2VvbV92bGluZSh4aW50ZXJjZXB0ID0gYygtMC4wNywgMC4wNyksIGxpbmV0eXBlID0gImRvdHRlZCIpICsNCiAgYXBhdGhlbWUgKw0KICB0aGVtZShsZWdlbmQucG9zaXRpb24gPSAibm9uZSIpICsNCiAgc2NhbGVfZmlsbF9tYW51YWwodmFsdWVzID0gYygiZ3JheTgwIiwgInNreWJsdWUiKSkgKw0KICBsYWJzKHggPSAiRWZmZWN0IFNpemUgRXN0aW1hdGVzIiwgeSA9ICJQcmVkaWN0b3JzIikgKw0KICB4bGltICgtMC42LCAwLjYpDQoNCg0KICANCmBgYA0KDQojIyMgUmVsYXRpb25zaGlwIFNhdGlzZmFjdGlvbiB7LnRhYnNldH0NCiMjIyMgTW9kZWwNCmBgYHtyfQ0KbV9jb25ncnVlbmN5X3JlbHNhdCA9IGJybShyZWxhdGlvbnNoaXBfc2F0aXNmYWN0aW9uIH4NCiAgICAgICAgICAgICAgICAgICAgICAgICAgY29udHJhY2VwdGlvbl9ob3Jtb25hbCAqIGNvbmdydWVudF9jb250cmFjZXB0aW9uLA0KICAgICAgICAgICAgICAgIGRhdGEgPSBkYXRhLCBmYW1pbHkgPSBnYXVzc2lhbigpLA0KICAgICAgICAgICAgICAgIGZpbGUgPSAibV9jb25ncnVlbmN5X3JlbHNhdCIpDQpgYGANCg0KIyMjIyBTdW1tYXJ5DQpgYGB7cn0NCnN1bW1hcnkobV9jb25ncnVlbmN5X3JlbHNhdCwgaW50ZXJ2YWxzID0gVCwgcHJvYiA9IDAuOTApDQpgYGANCg0KIyMjIyBDb21wYXJpc29uIHdpdGggUk9QRQ0KYGBge3J9DQpwbG90KGVxdWl2YWxlbmNlX3Rlc3QobV9jb25ncnVlbmN5X3JlbHNhdCwgcmFuZ2UgPSBjKC0wLjA0LCAwLjA0KSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpKQ0KZXF1aXZhbGVuY2VfdGVzdChtX2NvbmdydWVuY3lfcmVsc2F0LCByYW5nZSA9IGMoLTAuMDQsIDAuMDQpLCBjaSA9IDAuOTAsDQogICAgICAgICAgICAgICAgICAgICAgcGFyYW1ldGVycyA9ICJjb250cmFjZXB0aW9uIikNCmBgYA0KDQojIyMjIFBsb3RzDQpgYGB7ciB3YXJuaW5nID0gRkFMU0V9DQpjb25kaXRpb25hbF9lZmZlY3RzKG1fY29uZ3J1ZW5jeV9yZWxzYXQsIGFzayA9IEZBTFNFKQ0KYGBgDQoNCiMjIyMgRm9yZXN0IFBsb3QgZm9yIEVmZmVjdCBTaXplcyB7LmFjdGl2ZX0NCmBgYHtyfQ0KbV9jb25ncnVlbmN5X3JlbHNhdCAlPiUNCiAgc3ByZWFkX2RyYXdzKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcywgYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEsDQogICAgICAgICAgICAgICBgYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzOmNvbmdydWVudF9jb250cmFjZXB0aW9uMWApICU+JQ0KICBwaXZvdF9sb25nZXIoY29scyA9IGMoYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzLCBiX2NvbmdydWVudF9jb250cmFjZXB0aW9uMSwNCiAgICAgICAgICAgICAgIGBiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXM6Y29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xYCksDQogICAgICAgICAgICAgICBuYW1lc190byA9ICJjb25kaXRpb24iLA0KICAgICAgICAgICAgICAgdmFsdWVzX3RvID0gInJfY29uZGl0aW9uIikgJT4lDQogIG11dGF0ZShjb25kaXRpb25fbWVhbiA9IHJfY29uZGl0aW9uLA0KICAgICAgICAgZ3JvdXAgPSBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgIm9udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAiQ29udHJhY2VwdGlvbiIsIE5BKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGlmZWxzZShjb25kaXRpb24gPT0gImJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAiSG9ybW9uYWwgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbmdydWVudF9jb250cmFjZXB0aW9uMSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbmdydWVudCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllczpjb25ncnVlbnRfY29udHJhY2VwdGlvbjEiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICJJbnRlcmFjdGlvbiBIb3Jtb25hbCBDb250cmFjcGV0aW9uIGFuZCBDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgY29uZGl0aW9uKSkpLA0KICAgICAgICAgY29uZGl0aW9uID0gZmFjdG9yKGNvbmRpdGlvbiwgbGV2ZWxzID0gcmV2KGMoIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiQ29uZ3J1ZW50IENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiSW50ZXJhY3Rpb24gSG9ybW9uYWwgQ29udHJhY3BldGlvbiBhbmQgQ29uZ3J1ZW50IENvbnRyYWNlcHRpb24iKSkpKSAlPiUNCiAgZ2dwbG90KGFlcyh5ID0gY29uZGl0aW9uLA0KICAgICAgICAgICAgIHggPSBjb25kaXRpb25fbWVhbiwNCiAgICAgICAgICAgICBmaWxsID0gc3RhdChhYnMoeCkgPCAwLjA0KSkpICsNCiAgc3RhdF9oYWxmZXllKCkgKw0KICBnZW9tX3ZsaW5lKHhpbnRlcmNlcHQgPSBjKC0wLjA0LCAwLjA0KSwgbGluZXR5cGUgPSAiZG90dGVkIikgKw0KICBhcGF0aGVtZSArDQogIHRoZW1lKGxlZ2VuZC5wb3NpdGlvbiA9ICJub25lIikgKw0KICBzY2FsZV9maWxsX21hbnVhbCh2YWx1ZXMgPSBjKCJncmF5ODAiLCAic2t5Ymx1ZSIpKSArDQogIGxhYnMoeCA9ICJFZmZlY3QgU2l6ZSBFc3RpbWF0ZXMiLCB5ID0gIlByZWRpY3RvcnMiKSArDQogIHhsaW0gKC0wLjYsIDAuNikNCg0KYGBgDQoNCiMjIyBTZXh1YWwgU2F0aXNmYWN0aW9uIHsudGFic2V0fQ0KIyMjIyBNb2RlbA0KYGBge3J9DQptX2NvbmdydWVuY3lfc2V4c2F0ID0gYnJtKHNhdGlzZmFjdGlvbl9zZXh1YWxfaW50ZXJjb3Vyc2Ugfg0KICAgICAgICAgICAgICAgICAgICAgICAgICBjb250cmFjZXB0aW9uX2hvcm1vbmFsICogY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24sDQogICAgICAgICAgICAgICAgZGF0YSA9IGRhdGEsIGZhbWlseSA9IGdhdXNzaWFuKCksDQogICAgICAgICAgICAgICAgZmlsZSA9ICJtX2NvbmdydWVuY3lfc2V4c2F0IikNCmBgYA0KDQojIyMjIFN1bW1hcnkNCmBgYHtyfQ0Kc3VtbWFyeShtX2NvbmdydWVuY3lfc2V4c2F0LCBpbnRlcnZhbHMgPSBULCBwcm9iID0gMC45MCkNCmBgYA0KDQojIyMjIENvbXBhcmlzb24gd2l0aCBST1BFDQpgYGB7cn0NCnBsb3QoZXF1aXZhbGVuY2VfdGVzdChtX2NvbmdydWVuY3lfc2V4c2F0LCByYW5nZSA9IGMoLTAuMTEsIDAuMTEpLCBjaSA9IDAuOTAsDQogICAgICAgICAgICAgICAgICAgICAgcGFyYW1ldGVycyA9ICJjb250cmFjZXB0aW9uIikpDQplcXVpdmFsZW5jZV90ZXN0KG1fY29uZ3J1ZW5jeV9zZXhzYXQsIHJhbmdlID0gYygtMC4xMSwgMC4xMSksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKQ0KYGBgDQoNCiMjIyMgUGxvdHMNCmBgYHtyIHdhcm5pbmcgPSBGQUxTRX0NCmNvbmRpdGlvbmFsX2VmZmVjdHMobV9jb25ncnVlbmN5X3NleHNhdCwgYXNrID0gRkFMU0UpDQpgYGANCg0KIyMjIyBGb3Jlc3QgUGxvdCBmb3IgRWZmZWN0IFNpemVzIHsuYWN0aXZlfQ0KYGBge3J9DQptX2NvbmdydWVuY3lfc2V4c2F0ICU+JQ0KICBzcHJlYWRfZHJhd3MoYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzLCBiX2NvbmdydWVudF9jb250cmFjZXB0aW9uMSwNCiAgICAgICAgICAgICAgIGBiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXM6Y29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xYCkgJT4lDQogIHBpdm90X2xvbmdlcihjb2xzID0gYyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMsIGJfY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xLA0KICAgICAgICAgICAgICAgYGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllczpjb25ncnVlbnRfY29udHJhY2VwdGlvbjFgKSwNCiAgICAgICAgICAgICAgIG5hbWVzX3RvID0gImNvbmRpdGlvbiIsDQogICAgICAgICAgICAgICB2YWx1ZXNfdG8gPSAicl9jb25kaXRpb24iKSAlPiUNCiAgbXV0YXRlKGNvbmRpdGlvbl9tZWFuID0gcl9jb25kaXRpb24sDQogICAgICAgICBncm91cCA9IGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAib250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJDb250cmFjZXB0aW9uIiwgTkEpLA0KICAgICAgICAgY29uZGl0aW9uID0gaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJIb3Jtb25hbCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAiQ29uZ3J1ZW50IENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzOmNvbmdydWVudF9jb250cmFjZXB0aW9uMSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgIkludGVyYWN0aW9uIEhvcm1vbmFsIENvbnRyYWNwZXRpb24gYW5kIENvbmdydWVudCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICBjb25kaXRpb24pKSksDQogICAgICAgICBjb25kaXRpb24gPSBmYWN0b3IoY29uZGl0aW9uLCBsZXZlbHMgPSByZXYoYygiSG9ybW9uYWwgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJJbnRlcmFjdGlvbiBIb3Jtb25hbCBDb250cmFjcGV0aW9uIGFuZCBDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIpKSkpICU+JQ0KICBnZ3Bsb3QoYWVzKHkgPSBjb25kaXRpb24sDQogICAgICAgICAgICAgeCA9IGNvbmRpdGlvbl9tZWFuLA0KICAgICAgICAgICAgIGZpbGwgPSBzdGF0KGFicyh4KSA8IDAuMTEpKSkgKw0KICBzdGF0X2hhbGZleWUoKSArDQogIGdlb21fdmxpbmUoeGludGVyY2VwdCA9IGMoLTAuMTEsIDAuMTEpLCBsaW5ldHlwZSA9ICJkb3R0ZWQiKSArDQogIGFwYXRoZW1lICsNCiAgdGhlbWUobGVnZW5kLnBvc2l0aW9uID0gIm5vbmUiKSArDQogIHNjYWxlX2ZpbGxfbWFudWFsKHZhbHVlcyA9IGMoImdyYXk4MCIsICJza3libHVlIikpICsNCiAgbGFicyh4ID0gIkVmZmVjdCBTaXplIEVzdGltYXRlcyIsIHkgPSAiUHJlZGljdG9ycyIpICsNCiAgeGxpbSAoLTAuNiwgMC42KQ0KYGBgDQoNCiMjIyBMaWJpZG8gey50YWJzZXR9DQojIyMjIE1vZGVsDQpgYGB7cn0NCm1fY29uZ3J1ZW5jeV9saWJpZG8gPSBicm0oZGlhcnlfbGliaWRvX21lYW4gfg0KICAgICAgICAgICAgICAgICAgICAgICAgICBjb250cmFjZXB0aW9uX2hvcm1vbmFsICogY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24sDQogICAgICAgICAgICAgICAgZGF0YSA9IGRhdGEsIGZhbWlseSA9IGdhdXNzaWFuKCksDQogICAgICAgICAgICAgICAgZmlsZSA9ICJtX2NvbmdydWVuY3lfbGliaWRvIikNCg0KYGBgDQoNCiMjIyMgU3VtbWFyeQ0KYGBge3J9DQpzdW1tYXJ5KG1fY29uZ3J1ZW5jeV9saWJpZG8sIGludGVydmFscyA9IFQsIHByb2IgPSAwLjkwKQ0KYGBgDQoNCiMjIyMgQ29tcGFyaXNvbiB3aXRoIFJPUEUNCmBgYHtyfQ0KcGxvdChlcXVpdmFsZW5jZV90ZXN0KG1fY29uZ3J1ZW5jeV9saWJpZG8sIHJhbmdlID0gYygtMC4wNiwgMC4wNiksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKSkNCmVxdWl2YWxlbmNlX3Rlc3QobV9jb25ncnVlbmN5X2xpYmlkbywgcmFuZ2UgPSBjKC0wLjA2LCAwLjA2KSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpDQpgYGANCg0KIyMjIyBQbG90cw0KYGBge3Igd2FybmluZyA9IEZBTFNFfQ0KY29uZGl0aW9uYWxfZWZmZWN0cyhtX2NvbmdydWVuY3lfbGliaWRvLCBhc2sgPSBGQUxTRSkNCmBgYA0KDQojIyMjIEZvcmVzdCBQbG90IGZvciBFZmZlY3QgU2l6ZXMgey5hY3RpdmV9DQpgYGB7cn0NCm1fY29uZ3J1ZW5jeV9saWJpZG8gJT4lDQogIHNwcmVhZF9kcmF3cyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMsIGJfY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xLA0KICAgICAgICAgICAgICAgYGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllczpjb25ncnVlbnRfY29udHJhY2VwdGlvbjFgKSAlPiUNCiAgcGl2b3RfbG9uZ2VyKGNvbHMgPSBjKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcywgYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEsDQogICAgICAgICAgICAgICBgYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzOmNvbmdydWVudF9jb250cmFjZXB0aW9uMWApLA0KICAgICAgICAgICAgICAgbmFtZXNfdG8gPSAiY29uZGl0aW9uIiwNCiAgICAgICAgICAgICAgIHZhbHVlc190byA9ICJyX2NvbmRpdGlvbiIpICU+JQ0KICBtdXRhdGUoY29uZGl0aW9uX21lYW4gPSByX2NvbmRpdGlvbiwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnRyYWNlcHRpb24iLCBOQSksDQogICAgICAgICBjb25kaXRpb24gPSBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXM6Y29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAiSW50ZXJhY3Rpb24gSG9ybW9uYWwgQ29udHJhY3BldGlvbiBhbmQgQ29uZ3J1ZW50IENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgIGNvbmRpdGlvbikpKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGZhY3Rvcihjb25kaXRpb24sIGxldmVscyA9IHJldihjKCJIb3Jtb25hbCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbmdydWVudCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkludGVyYWN0aW9uIEhvcm1vbmFsIENvbnRyYWNwZXRpb24gYW5kIENvbmdydWVudCBDb250cmFjZXB0aW9uIikpKSkgJT4lDQogIGdncGxvdChhZXMoeSA9IGNvbmRpdGlvbiwNCiAgICAgICAgICAgICB4ID0gY29uZGl0aW9uX21lYW4sDQogICAgICAgICAgICAgZmlsbCA9IHN0YXQoYWJzKHgpIDwgMC4wNikpKSArDQogIHN0YXRfaGFsZmV5ZSgpICsNCiAgZ2VvbV92bGluZSh4aW50ZXJjZXB0ID0gYygtMC4wNiwgMC4wNiksIGxpbmV0eXBlID0gImRvdHRlZCIpICsNCiAgYXBhdGhlbWUgKw0KICB0aGVtZShsZWdlbmQucG9zaXRpb24gPSAibm9uZSIpICsNCiAgc2NhbGVfZmlsbF9tYW51YWwodmFsdWVzID0gYygiZ3JheTgwIiwgInNreWJsdWUiKSkgKw0KICBsYWJzKHggPSAiRWZmZWN0IFNpemUgRXN0aW1hdGVzIiwgeSA9ICJQcmVkaWN0b3JzIikgKw0KICB4bGltICgtMC42LCAwLjYpDQpgYGANCg0KDQojIyMgU2V4dWFsIEZyZXF1ZW5jeSAoUGVuZXRyYXRpdmUgSW50ZXJjb3Vyc2UpIHsudGFic2V0fQ0KIyMjIyBNb2RlbA0KYGBge3J9DQptX2NvbmdydWVuY3lfc2V4ZnJlcXBlbiA9IGJybShkaWFyeV9zZXhfYWN0aXZlX3NleF9zdW0gfiBvZmZzZXQobG9nKG51bWJlcl9vZl9kYXlzKSkgKw0KICAgICAgICAgICAgICAgICAgICAgY29udHJhY2VwdGlvbl9ob3Jtb25hbCAqIGNvbmdydWVudF9jb250cmFjZXB0aW9uLA0KICAgICAgICAgICAgICAgIGRhdGEgPSBkYXRhLCBmYW1pbHkgPSBwb2lzc29uKCksDQogICAgICAgICAgICAgICAgZmlsZSA9ICJtX2NvbmdydWVuY3lfc2V4ZnJlcXBlbiIpDQpgYGANCg0KIyMjIyBTdW1tYXJ5DQpgYGB7cn0NCnN1bW1hcnkobV9jb25ncnVlbmN5X3NleGZyZXFwZW4sIGludGVydmFscyA9IFQsIHByb2IgPSAwLjkwKQ0KYGBgDQoNCiMjIyMgQ29tcGFyaXNvbiB3aXRoIFJPUEUNCmBgYHtyfQ0KcGxvdChlcXVpdmFsZW5jZV90ZXN0KG1fY29uZ3J1ZW5jeV9zZXhmcmVxcGVuLCByYW5nZSA9IGMoLTAuMDUsIDAuMDUpLCBjaSA9IDAuOTAsDQogICAgICAgICAgICAgICAgICAgICAgcGFyYW1ldGVycyA9ICJjb250cmFjZXB0aW9uIikpDQplcXVpdmFsZW5jZV90ZXN0KG1fY29uZ3J1ZW5jeV9zZXhmcmVxcGVuLCByYW5nZSA9IGMoLTAuMDUsIDAuMDUpLCBjaSA9IDAuOTAsDQogICAgICAgICAgICAgICAgICAgICAgcGFyYW1ldGVycyA9ICJjb250cmFjZXB0aW9uIikNCmBgYA0KDQojIyMjIFBsb3RzDQpgYGB7ciB3YXJuaW5nID0gRkFMU0V9DQpjb25kaXRpb25hbF9lZmZlY3RzKG1fY29uZ3J1ZW5jeV9zZXhmcmVxcGVuLA0KICAgICAgICAgICAgICAgICAgICBlZmZlY3RzID0gImNvbnRyYWNlcHRpb25faG9ybW9uYWw6Y29uZ3J1ZW50X2NvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICBjb25kaXRpb25zID0gZGF0YS5mcmFtZShudW1iZXJfb2ZfZGF5cyA9IDEpKQ0KYGBgDQoNCiMjIyMgRm9yZXN0IFBsb3QgZm9yIEVmZmVjdCBTaXplcyB7LmFjdGl2ZX0NCmBgYHtyfQ0KbV9jb25ncnVlbmN5X3NleGZyZXFwZW4gJT4lDQogIHNwcmVhZF9kcmF3cyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMsIGJfY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xLA0KICAgICAgICAgICAgICAgYGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllczpjb25ncnVlbnRfY29udHJhY2VwdGlvbjFgKSAlPiUNCiAgcGl2b3RfbG9uZ2VyKGNvbHMgPSBjKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcywgYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEsDQogICAgICAgICAgICAgICBgYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzOmNvbmdydWVudF9jb250cmFjZXB0aW9uMWApLA0KICAgICAgICAgICAgICAgbmFtZXNfdG8gPSAiY29uZGl0aW9uIiwNCiAgICAgICAgICAgICAgIHZhbHVlc190byA9ICJyX2NvbmRpdGlvbiIpICU+JQ0KICBtdXRhdGUoY29uZGl0aW9uX21lYW4gPSByX2NvbmRpdGlvbiwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnRyYWNlcHRpb24iLCBOQSksDQogICAgICAgICBjb25kaXRpb24gPSBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXM6Y29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAiSW50ZXJhY3Rpb24gSG9ybW9uYWwgQ29udHJhY3BldGlvbiBhbmQgQ29uZ3J1ZW50IENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgIGNvbmRpdGlvbikpKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGZhY3Rvcihjb25kaXRpb24sIGxldmVscyA9IHJldihjKCJIb3Jtb25hbCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbmdydWVudCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkludGVyYWN0aW9uIEhvcm1vbmFsIENvbnRyYWNwZXRpb24gYW5kIENvbmdydWVudCBDb250cmFjZXB0aW9uIikpKSkgJT4lDQogIGdncGxvdChhZXMoeSA9IGNvbmRpdGlvbiwNCiAgICAgICAgICAgICB4ID0gY29uZGl0aW9uX21lYW4sDQogICAgICAgICAgICAgZmlsbCA9IHN0YXQoYWJzKHgpIDwgMC4wNSkpKSArDQogIHN0YXRfaGFsZmV5ZSgpICsNCiAgZ2VvbV92bGluZSh4aW50ZXJjZXB0ID0gYygtMC4wNSwgMC4wNTEpLCBsaW5ldHlwZSA9ICJkb3R0ZWQiKSArDQogIGFwYXRoZW1lICsNCiAgdGhlbWUobGVnZW5kLnBvc2l0aW9uID0gIm5vbmUiKSArDQogIHNjYWxlX2ZpbGxfbWFudWFsKHZhbHVlcyA9IGMoImdyYXk4MCIsICJza3libHVlIikpICsNCiAgbGFicyh4ID0gIkVmZmVjdCBTaXplIEVzdGltYXRlcyIsIHkgPSAiUHJlZGljdG9ycyIpDQpgYGANCg0KIyMjIE1hc3R1cmJhdGlvbiBGcmVxdWVuY3kgey50YWJzZXR9DQojIyMjIE1vZGVsDQpgYGB7cn0NCm1fY29uZ3J1ZW5jeV9tYXNmcmVxID0gYnJtKGRpYXJ5X21hc3R1cmJhdGlvbl9zdW0gfiBvZmZzZXQobG9nKG51bWJlcl9vZl9kYXlzKSkgKw0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICBjb250cmFjZXB0aW9uX2hvcm1vbmFsICogY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24sDQogICAgICAgICAgICAgICAgZGF0YSA9IGRhdGEsIGZhbWlseSA9IHBvaXNzb24oKSwNCiAgICAgICAgICAgICAgICBmaWxlID0gIm1fY29uZ3J1ZW5jeV9tYXNmcmVxIikNCmBgYA0KDQojIyMjIFN1bW1hcnkNCmBgYHtyfQ0Kc3VtbWFyeShtX2NvbmdydWVuY3lfbWFzZnJlcSwgaW50ZXJ2YWxzID0gVCwgcHJvYiA9IDAuOTApDQpgYGANCg0KIyMjIyBDb21wYXJpc29uIHdpdGggUk9QRQ0KYGBge3J9DQpwbG90KGVxdWl2YWxlbmNlX3Rlc3QobV9jb25ncnVlbmN5X21hc2ZyZXEsIHJhbmdlID0gYygtMC4wNSwgMC4wNSksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKSkNCmVxdWl2YWxlbmNlX3Rlc3QobV9jb25ncnVlbmN5X21hc2ZyZXEsIHJhbmdlID0gYygtMC4wNSwgMC4wNSksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKQ0KYGBgDQoNCiMjIyMgUGxvdHMNCmBgYHtyIHdhcm5pbmcgPSBGQUxTRX0NCmNvbmRpdGlvbmFsX2VmZmVjdHMobV9jb25ncnVlbmN5X21hc2ZyZXEsDQogICAgICAgICAgICAgICAgICAgIGVmZmVjdHMgPSAiY29udHJhY2VwdGlvbl9ob3Jtb25hbDpjb25ncnVlbnRfY29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgIGNvbmRpdGlvbnMgPSBkYXRhLmZyYW1lKG51bWJlcl9vZl9kYXlzID0gMSkpDQpgYGANCg0KIyMjIyBGb3Jlc3QgUGxvdCBmb3IgRWZmZWN0IFNpemVzIHsuYWN0aXZlfQ0KYGBge3J9DQptX2NvbmdydWVuY3lfbWFzZnJlcSAlPiUNCnNwcmVhZF9kcmF3cyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMsIGJfY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xLA0KICAgICAgICAgICAgICAgYGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllczpjb25ncnVlbnRfY29udHJhY2VwdGlvbjFgKSAlPiUNCiAgcGl2b3RfbG9uZ2VyKGNvbHMgPSBjKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcywgYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEsDQogICAgICAgICAgICAgICBgYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzOmNvbmdydWVudF9jb250cmFjZXB0aW9uMWApLA0KICAgICAgICAgICAgICAgbmFtZXNfdG8gPSAiY29uZGl0aW9uIiwNCiAgICAgICAgICAgICAgIHZhbHVlc190byA9ICJyX2NvbmRpdGlvbiIpICU+JQ0KICBtdXRhdGUoY29uZGl0aW9uX21lYW4gPSByX2NvbmRpdGlvbiwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnRyYWNlcHRpb24iLCBOQSksDQogICAgICAgICBjb25kaXRpb24gPSBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXM6Y29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAiSW50ZXJhY3Rpb24gSG9ybW9uYWwgQ29udHJhY3BldGlvbiBhbmQgQ29uZ3J1ZW50IENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgIGNvbmRpdGlvbikpKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGZhY3Rvcihjb25kaXRpb24sIGxldmVscyA9IHJldihjKCJIb3Jtb25hbCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbmdydWVudCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkludGVyYWN0aW9uIEhvcm1vbmFsIENvbnRyYWNwZXRpb24gYW5kIENvbmdydWVudCBDb250cmFjZXB0aW9uIikpKSkgJT4lDQogIGdncGxvdChhZXMoeSA9IGNvbmRpdGlvbiwNCiAgICAgICAgICAgICB4ID0gY29uZGl0aW9uX21lYW4sDQogICAgICAgICAgICAgZmlsbCA9IHN0YXQoYWJzKHgpIDwgMC4wNSkpKSArDQogIHN0YXRfaGFsZmV5ZSgpICsNCiAgZ2VvbV92bGluZSh4aW50ZXJjZXB0ID0gYygtMC4wNSwgMC4wNSksIGxpbmV0eXBlID0gImRvdHRlZCIpICsNCiAgYXBhdGhlbWUgKw0KICB0aGVtZShsZWdlbmQucG9zaXRpb24gPSAibm9uZSIpICsNCiAgc2NhbGVfZmlsbF9tYW51YWwodmFsdWVzID0gYygiZ3JheTgwIiwgInNreWJsdWUiKSkgKw0KICBsYWJzKHggPSAiRWZmZWN0IFNpemUgRXN0aW1hdGVzIiwgeSA9ICJQcmVkaWN0b3JzIikNCmBgYA0KDQoNCg0KIyMgQ29udHJvbGxlZCBNb2RlbHM6IEVmZmVjdHMgb2YgKEluKUNvbmdydWVuY3QgSEMgVXNlIHsudGFic2V0fQ0KIyMjIEF0dHJhY3RpdmVuZXNzIG9mIFBhcnRuZXIgIHsudGFic2V0fQ0KIyMjIyBNb2RlbA0KYGBge3J9DQptX2NvbmdydWVuY3lfYXRycl9jb250cm9sbGVkID0gYnJtKGF0dHJhY3RpdmVuZXNzX3BhcnRuZXIgfg0KICAgICAgICAgICAgICAgICAgICAgICAgICBjb250cmFjZXB0aW9uX2hvcm1vbmFsICogY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24gKw0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICBhZ2UgKyBuZXRfaW5jb21lICsgcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvciArDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICBlZHVjYXRpb25feWVhcnMgKw0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgYmZpX2V4dHJhICsgYmZpX25ldXJvICsgYmZpX2FncmVlICsgYmZpX2NvbnNjICsgYmZpX29wZW4gKw0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgcmVsaWdpb3NpdHksDQogICAgICAgICAgICAgICAgZGF0YSA9IGRhdGEsIGZhbWlseSA9IGdhdXNzaWFuKCksDQogICAgICAgICAgICAgICAgZmlsZSA9ICJtX2NvbmdydWVuY3lfYXRycl9jb250cm9sbGVkIikNCmBgYA0KDQojIyMjIFN1bW1hcnkNCmBgYHtyfQ0Kc3VtbWFyeShtX2NvbmdydWVuY3lfYXRycl9jb250cm9sbGVkLCBpbnRlcnZhbHMgPSBULCBwcm9iID0gMC45MCkNCmBgYA0KDQojIyMjIENvbXBhcmlzb24gd2l0aCBST1BFDQpgYGB7cn0NCnBsb3QoZXF1aXZhbGVuY2VfdGVzdChtX2NvbmdydWVuY3lfYXRycl9jb250cm9sbGVkLCByYW5nZSA9IGMoLTAuMDcsIDAuMDcpLCBjaSA9IDAuOTAsDQogICAgICAgICAgICAgICAgICAgICAgcGFyYW1ldGVycyA9ICJjb250cmFjZXB0aW9uIikpDQplcXVpdmFsZW5jZV90ZXN0KG1fY29uZ3J1ZW5jeV9hdHJyX2NvbnRyb2xsZWQsIHJhbmdlID0gYygtMC4wNywgMC4wNyksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKQ0KYGBgDQoNCiMjIyMgUGxvdHMNCmBgYHtyIHdhcm5pbmcgPSBGQUxTRX0NCmNvbmRpdGlvbmFsX2VmZmVjdHMobV9jb25ncnVlbmN5X2F0cnJfY29udHJvbGxlZCwgYXNrID0gRkFMU0UpDQpgYGANCg0KIyMjIyBGb3Jlc3QgUGxvdCBmb3IgRWZmZWN0IFNpemVzIHsuYWN0aXZlfQ0KYGBge3J9DQptX2NvbmdydWVuY3lfYXRycl9jb250cm9sbGVkICU+JQ0KICBzcHJlYWRfZHJhd3MoYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzLCBiX2NvbmdydWVudF9jb250cmFjZXB0aW9uMSwNCiAgICAgICAgICAgICAgIGBiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXM6Y29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xYCwNCiAgICAgICAgICAgICAgIGJfYWdlLA0KICAgICAgICAgICAgICAgYl9uZXRfaW5jb21lZXVyb181MDBfMTAwMCwgYl9uZXRfaW5jb21lZXVyb18xMDAwXzIwMDAsIA0KICAgICAgICAgICAgICAgYl9uZXRfaW5jb21lZXVyb18yMDAwXzMwMDAsIGJfbmV0X2luY29tZWV1cm9fZ3RfMzAwMCwgYl9uZXRfaW5jb21lZG9udF90ZWxsLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG8yOG1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvNTJtb250aHMsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfbW9yZXRoYW41Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfZWR1Y2F0aW9uX3llYXJzLA0KICAgICAgICAgICAgICAgYl9iZmlfZXh0cmEsIGJfYmZpX25ldXJvLCBiX2JmaV9hZ3JlZSwgYl9iZmlfY29uc2MsIGJfYmZpX29wZW4sIA0KICAgICAgICAgICAgICAgYl9yZWxpZ2lvc2l0eSkgJT4lDQogIHBpdm90X2xvbmdlcihjb2xzID0gYyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMsIGJfY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xLA0KICAgICAgICAgICAgICAgYGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllczpjb25ncnVlbnRfY29udHJhY2VwdGlvbjFgLA0KICAgICAgICAgICAgICAgICAgICAgICAgYl9hZ2UsDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzUwMF8xMDAwLCBiX25ldF9pbmNvbWVldXJvXzEwMDBfMjAwMCwgDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzIwMDBfMzAwMCwgYl9uZXRfaW5jb21lZXVyb19ndF8zMDAwLCBiX25ldF9pbmNvbWVkb250X3RlbGwsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzI4bW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG81Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF9tb3JldGhhbjUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9lZHVjYXRpb25feWVhcnMsDQogICAgICAgICAgICAgICBiX2JmaV9leHRyYSwgYl9iZmlfbmV1cm8sIGJfYmZpX2FncmVlLCBiX2JmaV9jb25zYywgYl9iZmlfb3BlbiwgDQogICAgICAgICAgICAgICBiX3JlbGlnaW9zaXR5KSwNCiAgICAgICAgICAgICAgIG5hbWVzX3RvID0gImNvbmRpdGlvbiIsDQogICAgICAgICAgICAgICB2YWx1ZXNfdG8gPSAicl9jb25kaXRpb24iKSAlPiUNCiAgbXV0YXRlKGNvbmRpdGlvbl9tZWFuID0gcl9jb25kaXRpb24sDQogICAgICAgICBncm91cCA9IGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJSZWxhdGlvbnNoaXAgRHVyYXRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX25ldF9pbmNvbWUiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJJbmNvbWUiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIE5BKSksDQogICAgICAgICBncm91cCA9IGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAib250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJDb250cmFjZXB0aW9uIiwgZ3JvdXApLA0KICAgICAgICAgY29uZGl0aW9uID0gaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJIb3Jtb25hbCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAiQ29uZ3J1ZW50IENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzOmNvbmdydWVudF9jb250cmFjZXB0aW9uMSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgIkludGVyYWN0aW9uIEhvcm1vbmFsIENvbnRyYWNwZXRpb24gYW5kIENvbmdydWVudCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICBjb25kaXRpb24pKSksDQogICAgICAgICBjb25kaXRpb24gPSBpZmVsc2UoY29uZGl0aW9uID09ICJiX2FnZSIsICJBZ2UiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fNTAwXzEwMDAiLCAiNTAwLTEwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb18xMDAwXzIwMDAiLCAiMTAwMC0yMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fMjAwMF8zMDAwIiwgIjIwMDAtMzAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVldXJvX2d0XzMwMDAiLCAiPjMwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZG9udF90ZWxsIiwgImRvIG5vdCB0ZWxsIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzI4bW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIjEzLTI4IG1vbnRocyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG81Mm1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICIyOS01MiBtb250aHMiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF9tb3JldGhhbjUybW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIj41MiBtb250aHMiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfZWR1Y2F0aW9uX3llYXJzIiwgIlllYXJzIG9mIEVkdWNhdGlvbiIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfZXh0cmEiLCAiRXh0cmF2ZXJzaW9uIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9uZXVybyIsICJOZXVyb3RpY2lzbSIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfYWdyZWUiLCAiQWdyZWVhYmxlbmVzcyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfY29uc2MiLCAiQ29uc2NpZW50aW91c25lc3MiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX29wZW4iLCAiT3Blbm5lc3MiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsaWdpb3NpdHkiLCAiUmVsaWdpb3NpdHkiLA0KICAgICAgICAgICAgICAgICAgICAgICBjb25kaXRpb24pKSkpKSkpKSkpKSkpKSkpLA0KICAgICAgICAgZ3JvdXAgPSBpZmVsc2UoaXMubmEoZ3JvdXApLCBjb25kaXRpb24sIGdyb3VwKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGZhY3Rvcihjb25kaXRpb24sIGxldmVscyA9IHJldihjKCJIb3Jtb25hbCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiQ29uZ3J1ZW50IENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJJbnRlcmFjdGlvbiBIb3Jtb25hbCBDb250cmFjcGV0aW9uIGFuZCBDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkFnZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIjUwMC0xMDAwIEV1cm8iLCAiMTAwMC0yMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICIyMDAwLTMwMDAgRXVybyIsICI+MzAwMCBFdXJvIiwgImRvIG5vdCB0ZWxsIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiMTMtMjggbW9udGhzIiwgIjI5LTUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIj41MiBtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJZZWFycyBvZiBFZHVjYXRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJFeHRyYXZlcnNpb24iLCAiTmV1cm90aWNpc20iLCAiQWdyZWVhYmxlbmVzcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25zY2llbnRpb3VzbmVzcyIsIk9wZW5uZXNzIiwiUmVsaWdpb3NpdHkiKSkpLA0KICAgICAgICAgZ3JvdXAgPSBmYWN0b3IoZ3JvdXAsIGxldmVscyA9IGMoIkNvbnRyYWNlcHRpb24iLCAiQWdlIiwgIkluY29tZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiUmVsYXRpb25zaGlwIER1cmF0aW9uIiwiWWVhcnMgb2YgRWR1Y2F0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJFeHRyYXZlcnNpb24iLCAiTmV1cm90aWNpc20iLCAiQWdyZWVhYmxlbmVzcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25zY2llbnRpb3VzbmVzcyIsIk9wZW5uZXNzIiwiUmVsaWdpb3NpdHkiKSkpICU+JQ0KICBnZ3Bsb3QoYWVzKHkgPSBjb25kaXRpb24sDQogICAgICAgICAgICAgeCA9IGNvbmRpdGlvbl9tZWFuLA0KICAgICAgICAgICAgIGZpbGwgPSBzdGF0KGFicyh4KSA8IDAuMDcpKSkgKw0KICBzdGF0X2hhbGZleWUoKSArDQogIGdlb21fdmxpbmUoeGludGVyY2VwdCA9IGMoLTAuMDcsIDAuMDcpLCBsaW5ldHlwZSA9ICJkb3R0ZWQiKSArDQogIGFwYXRoZW1lICsNCiAgdGhlbWUobGVnZW5kLnBvc2l0aW9uID0gIm5vbmUiKSArDQogIHNjYWxlX2ZpbGxfbWFudWFsKHZhbHVlcyA9IGMoImdyYXk4MCIsICJza3libHVlIikpICsNCiAgbGFicyh4ID0gIkVmZmVjdCBTaXplIEVzdGltYXRlcyIsIHkgPSAiUHJlZGljdG9ycyIpDQpgYGANCg0KIyMjIFJlbGF0aW9uc2hpcCBTYXRpc2ZhY3Rpb24gey50YWJzZXR9DQojIyMjIE1vZGVsDQpgYGB7cn0NCm1fY29uZ3J1ZW5jeV9yZWxzYXRfY29udHJvbGxlZCA9IGJybShyZWxhdGlvbnNoaXBfc2F0aXNmYWN0aW9uIH4NCiAgICAgICAgICAgICAgICAgICAgICAgICAgY29udHJhY2VwdGlvbl9ob3Jtb25hbCAqIGNvbmdydWVudF9jb250cmFjZXB0aW9uICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgYWdlICsgbmV0X2luY29tZSArIHJlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3IgKw0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgZWR1Y2F0aW9uX3llYXJzICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIGJmaV9leHRyYSArIGJmaV9uZXVybyArIGJmaV9hZ3JlZSArIGJmaV9jb25zYyArIGJmaV9vcGVuICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIHJlbGlnaW9zaXR5LA0KICAgICAgICAgICAgICAgIGRhdGEgPSBkYXRhLCBmYW1pbHkgPSBnYXVzc2lhbigpLA0KICAgICAgICAgICAgICAgIGZpbGUgPSAibV9jb25ncnVlbmN5X3JlbHNhdF9jb250cm9sbGVkIikNCmBgYA0KDQojIyMjIFN1bW1hcnkNCmBgYHtyfQ0Kc3VtbWFyeShtX2NvbmdydWVuY3lfcmVsc2F0X2NvbnRyb2xsZWQsIGludGVydmFscyA9IFQsIHByb2IgPSAwLjkwKQ0KYGBgDQoNCiMjIyMgQ29tcGFyaXNvbiB3aXRoIFJPUEUNCmBgYHtyfQ0KcGxvdChlcXVpdmFsZW5jZV90ZXN0KG1fY29uZ3J1ZW5jeV9yZWxzYXRfY29udHJvbGxlZCwgcmFuZ2UgPSBjKC0wLjA0LCAwLjA0KSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpKQ0KZXF1aXZhbGVuY2VfdGVzdChtX2NvbmdydWVuY3lfcmVsc2F0X2NvbnRyb2xsZWQsIHJhbmdlID0gYygtMC4wNCwgMC4wNCksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKQ0KYGBgDQoNCiMjIyMgUGxvdHMNCmBgYHtyIHdhcm5pbmcgPSBGQUxTRX0NCmNvbmRpdGlvbmFsX2VmZmVjdHMobV9jb25ncnVlbmN5X3JlbHNhdF9jb250cm9sbGVkLCBhc2sgPSBGQUxTRSkNCmBgYA0KDQojIyMjIEZvcmVzdCBQbG90IGZvciBFZmZlY3QgU2l6ZXMgey5hY3RpdmV9DQpgYGB7cn0NCm1fY29uZ3J1ZW5jeV9yZWxzYXRfY29udHJvbGxlZCAlPiUNCiAgDQogIHNwcmVhZF9kcmF3cyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMsIGJfY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xLA0KICAgICAgICAgICAgICAgYGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllczpjb25ncnVlbnRfY29udHJhY2VwdGlvbjFgLA0KICAgICAgICAgICAgICAgYl9hZ2UsDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzUwMF8xMDAwLCBiX25ldF9pbmNvbWVldXJvXzEwMDBfMjAwMCwgDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzIwMDBfMzAwMCwgYl9uZXRfaW5jb21lZXVyb19ndF8zMDAwLCBiX25ldF9pbmNvbWVkb250X3RlbGwsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzI4bW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG81Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF9tb3JldGhhbjUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9lZHVjYXRpb25feWVhcnMsDQogICAgICAgICAgICAgICBiX2JmaV9leHRyYSwgYl9iZmlfbmV1cm8sIGJfYmZpX2FncmVlLCBiX2JmaV9jb25zYywgYl9iZmlfb3BlbiwgDQogICAgICAgICAgICAgICBiX3JlbGlnaW9zaXR5KSAlPiUNCiAgcGl2b3RfbG9uZ2VyKGNvbHMgPSBjKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcywgYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEsDQogICAgICAgICAgICAgICBgYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzOmNvbmdydWVudF9jb250cmFjZXB0aW9uMWAsDQogICAgICAgICAgICAgICAgICAgICAgICBiX2FnZSwNCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fNTAwXzEwMDAsIGJfbmV0X2luY29tZWV1cm9fMTAwMF8yMDAwLCANCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fMjAwMF8zMDAwLCBiX25ldF9pbmNvbWVldXJvX2d0XzMwMDAsIGJfbmV0X2luY29tZWRvbnRfdGVsbCwNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvMjhtb250aHMsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX21vcmV0aGFuNTJtb250aHMsDQogICAgICAgICAgICAgICBiX2VkdWNhdGlvbl95ZWFycywNCiAgICAgICAgICAgICAgIGJfYmZpX2V4dHJhLCBiX2JmaV9uZXVybywgYl9iZmlfYWdyZWUsIGJfYmZpX2NvbnNjLCBiX2JmaV9vcGVuLCANCiAgICAgICAgICAgICAgIGJfcmVsaWdpb3NpdHkpLA0KICAgICAgICAgICAgICAgbmFtZXNfdG8gPSAiY29uZGl0aW9uIiwNCiAgICAgICAgICAgICAgIHZhbHVlc190byA9ICJyX2NvbmRpdGlvbiIpICU+JQ0KICBtdXRhdGUoY29uZGl0aW9uX21lYW4gPSByX2NvbmRpdGlvbiwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3IiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIlJlbGF0aW9uc2hpcCBEdXJhdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfbmV0X2luY29tZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkluY29tZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgTkEpKSwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnRyYWNlcHRpb24iLCBncm91cCksDQogICAgICAgICBjb25kaXRpb24gPSBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXM6Y29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAiSW50ZXJhY3Rpb24gSG9ybW9uYWwgQ29udHJhY3BldGlvbiBhbmQgQ29uZ3J1ZW50IENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgIGNvbmRpdGlvbikpKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGlmZWxzZShjb25kaXRpb24gPT0gImJfYWdlIiwgIkFnZSIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb181MDBfMTAwMCIsICI1MDAtMTAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVldXJvXzEwMDBfMjAwMCIsICIxMDAwLTIwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb18yMDAwXzMwMDAiLCAiMjAwMC0zMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fZ3RfMzAwMCIsICI+MzAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVkb250X3RlbGwiLCAiZG8gbm90IHRlbGwiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvMjhtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAiMTMtMjggbW9udGhzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzUybW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIjI5LTUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX21vcmV0aGFuNTJtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAiPjUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9lZHVjYXRpb25feWVhcnMiLCAiWWVhcnMgb2YgRWR1Y2F0aW9uIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9leHRyYSIsICJFeHRyYXZlcnNpb24iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX25ldXJvIiwgIk5ldXJvdGljaXNtIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9hZ3JlZSIsICJBZ3JlZWFibGVuZXNzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9jb25zYyIsICJDb25zY2llbnRpb3VzbmVzcyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfb3BlbiIsICJPcGVubmVzcyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxpZ2lvc2l0eSIsICJSZWxpZ2lvc2l0eSIsDQogICAgICAgICAgICAgICAgICAgICAgIGNvbmRpdGlvbikpKSkpKSkpKSkpKSkpKSksDQogICAgICAgICBncm91cCA9IGlmZWxzZShpcy5uYShncm91cCksIGNvbmRpdGlvbiwgZ3JvdXApLA0KICAgICAgICAgY29uZGl0aW9uID0gZmFjdG9yKGNvbmRpdGlvbiwgbGV2ZWxzID0gcmV2KGMoIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkludGVyYWN0aW9uIEhvcm1vbmFsIENvbnRyYWNwZXRpb24gYW5kIENvbmdydWVudCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiQWdlIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiNTAwLTEwMDAgRXVybyIsICIxMDAwLTIwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIjIwMDAtMzAwMCBFdXJvIiwgIj4zMDAwIEV1cm8iLCAiZG8gbm90IHRlbGwiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICIxMy0yOCBtb250aHMiLCAiMjktNTIgbW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiPjUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIlllYXJzIG9mIEVkdWNhdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkV4dHJhdmVyc2lvbiIsICJOZXVyb3RpY2lzbSIsICJBZ3JlZWFibGVuZXNzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnNjaWVudGlvdXNuZXNzIiwiT3Blbm5lc3MiLCJSZWxpZ2lvc2l0eSIpKSksDQogICAgICAgICBncm91cCA9IGZhY3Rvcihncm91cCwgbGV2ZWxzID0gYygiQ29udHJhY2VwdGlvbiIsICJBZ2UiLCAiSW5jb21lIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJSZWxhdGlvbnNoaXAgRHVyYXRpb24iLCJZZWFycyBvZiBFZHVjYXRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkV4dHJhdmVyc2lvbiIsICJOZXVyb3RpY2lzbSIsICJBZ3JlZWFibGVuZXNzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnNjaWVudGlvdXNuZXNzIiwiT3Blbm5lc3MiLCJSZWxpZ2lvc2l0eSIpKSkgJT4lDQogIGdncGxvdChhZXMoeSA9IGNvbmRpdGlvbiwNCiAgICAgICAgICAgICB4ID0gY29uZGl0aW9uX21lYW4sDQogICAgICAgICAgICAgZmlsbCA9IHN0YXQoYWJzKHgpIDwgMC4wNCkpKSArDQogIHN0YXRfaGFsZmV5ZSgpICsNCiAgZ2VvbV92bGluZSh4aW50ZXJjZXB0ID0gYygtMC4wNCwgMC4wNCksIGxpbmV0eXBlID0gImRvdHRlZCIpICsNCiAgYXBhdGhlbWUgKw0KICB0aGVtZShsZWdlbmQucG9zaXRpb24gPSAibm9uZSIpICsNCiAgc2NhbGVfZmlsbF9tYW51YWwodmFsdWVzID0gYygiZ3JheTgwIiwgInNreWJsdWUiKSkgKw0KICBsYWJzKHggPSAiRWZmZWN0IFNpemUgRXN0aW1hdGVzIiwgeSA9ICJQcmVkaWN0b3JzIikNCmBgYA0KDQojIyMgU2V4dWFsIFNhdGlzZmFjdGlvbiB7LnRhYnNldH0NCiMjIyMgTW9kZWwNCmBgYHtyfQ0KbV9jb25ncnVlbmN5X3NleHNhdF9jb250cm9sbGVkID0gYnJtKHNhdGlzZmFjdGlvbl9zZXh1YWxfaW50ZXJjb3Vyc2Ugfg0KICAgICAgICAgICAgICAgICAgICAgICAgICBjb250cmFjZXB0aW9uX2hvcm1vbmFsICogY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24gKw0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICBhZ2UgKyBuZXRfaW5jb21lICsgcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvciArDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICBlZHVjYXRpb25feWVhcnMgKw0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgYmZpX2V4dHJhICsgYmZpX25ldXJvICsgYmZpX2FncmVlICsgYmZpX2NvbnNjICsgYmZpX29wZW4gKw0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgcmVsaWdpb3NpdHksDQogICAgICAgICAgICAgICAgZGF0YSA9IGRhdGEsIGZhbWlseSA9IGdhdXNzaWFuKCksDQogICAgICAgICAgICAgICAgZmlsZSA9ICJtX2NvbmdydWVuY3lfc2V4c2F0X2NvbnRyb2xsZWQiKQ0KYGBgDQoNCiMjIyMgU3VtbWFyeQ0KYGBge3J9DQpzdW1tYXJ5KG1fY29uZ3J1ZW5jeV9zZXhzYXRfY29udHJvbGxlZCwgaW50ZXJ2YWxzID0gVCwgcHJvYiA9IDAuOTApDQpgYGANCg0KIyMjIyBDb21wYXJpc29uIHdpdGggUk9QRQ0KYGBge3J9DQpwbG90KGVxdWl2YWxlbmNlX3Rlc3QobV9jb25ncnVlbmN5X3NleHNhdF9jb250cm9sbGVkLCByYW5nZSA9IGMoLTAuMTEsIDAuMTEpLCBjaSA9IDAuOTAsDQogICAgICAgICAgICAgICAgICAgICAgcGFyYW1ldGVycyA9ICJjb250cmFjZXB0aW9uIikpDQplcXVpdmFsZW5jZV90ZXN0KG1fY29uZ3J1ZW5jeV9zZXhzYXRfY29udHJvbGxlZCwgcmFuZ2UgPSBjKC0wLjExLCAwLjExKSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpDQpgYGANCg0KIyMjIyBQbG90cw0KYGBge3Igd2FybmluZyA9IEZBTFNFfQ0KY29uZGl0aW9uYWxfZWZmZWN0cyhtX2NvbmdydWVuY3lfc2V4c2F0X2NvbnRyb2xsZWQsIGFzayA9IEZBTFNFKQ0KYGBgDQoNCiMjIyMgRm9yZXN0IFBsb3QgZm9yIEVmZmVjdCBTaXplcyB7LmFjdGl2ZX0NCmBgYHtyfQ0KbV9jb25ncnVlbmN5X3NleHNhdF9jb250cm9sbGVkICU+JQ0KICANCiAgc3ByZWFkX2RyYXdzKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcywgYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEsDQogICAgICAgICAgICAgICBgYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzOmNvbmdydWVudF9jb250cmFjZXB0aW9uMWAsDQogICAgICAgICAgICAgICBiX2FnZSwNCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fNTAwXzEwMDAsIGJfbmV0X2luY29tZWV1cm9fMTAwMF8yMDAwLCANCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fMjAwMF8zMDAwLCBiX25ldF9pbmNvbWVldXJvX2d0XzMwMDAsIGJfbmV0X2luY29tZWRvbnRfdGVsbCwNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvMjhtb250aHMsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX21vcmV0aGFuNTJtb250aHMsDQogICAgICAgICAgICAgICBiX2VkdWNhdGlvbl95ZWFycywNCiAgICAgICAgICAgICAgIGJfYmZpX2V4dHJhLCBiX2JmaV9uZXVybywgYl9iZmlfYWdyZWUsIGJfYmZpX2NvbnNjLCBiX2JmaV9vcGVuLCANCiAgICAgICAgICAgICAgIGJfcmVsaWdpb3NpdHkpICU+JQ0KICBwaXZvdF9sb25nZXIoY29scyA9IGMoYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzLCBiX2NvbmdydWVudF9jb250cmFjZXB0aW9uMSwNCiAgICAgICAgICAgICAgIGBiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXM6Y29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xYCwNCiAgICAgICAgICAgICAgICAgICAgICAgIGJfYWdlLA0KICAgICAgICAgICAgICAgYl9uZXRfaW5jb21lZXVyb181MDBfMTAwMCwgYl9uZXRfaW5jb21lZXVyb18xMDAwXzIwMDAsIA0KICAgICAgICAgICAgICAgYl9uZXRfaW5jb21lZXVyb18yMDAwXzMwMDAsIGJfbmV0X2luY29tZWV1cm9fZ3RfMzAwMCwgYl9uZXRfaW5jb21lZG9udF90ZWxsLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG8yOG1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvNTJtb250aHMsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfbW9yZXRoYW41Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfZWR1Y2F0aW9uX3llYXJzLA0KICAgICAgICAgICAgICAgYl9iZmlfZXh0cmEsIGJfYmZpX25ldXJvLCBiX2JmaV9hZ3JlZSwgYl9iZmlfY29uc2MsIGJfYmZpX29wZW4sIA0KICAgICAgICAgICAgICAgYl9yZWxpZ2lvc2l0eSksDQogICAgICAgICAgICAgICBuYW1lc190byA9ICJjb25kaXRpb24iLA0KICAgICAgICAgICAgICAgdmFsdWVzX3RvID0gInJfY29uZGl0aW9uIikgJT4lDQogIG11dGF0ZShjb25kaXRpb25fbWVhbiA9IHJfY29uZGl0aW9uLA0KICAgICAgICAgZ3JvdXAgPSBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvciIsDQogICAgICAgICAgICAgICAgICAgICAgICAiUmVsYXRpb25zaGlwIER1cmF0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAiYl9uZXRfaW5jb21lIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiSW5jb21lIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICBOQSkpLA0KICAgICAgICAgZ3JvdXAgPSBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgIm9udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAiQ29udHJhY2VwdGlvbiIsIGdyb3VwKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGlmZWxzZShjb25kaXRpb24gPT0gImJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAiSG9ybW9uYWwgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbmdydWVudF9jb250cmFjZXB0aW9uMSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbmdydWVudCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllczpjb25ncnVlbnRfY29udHJhY2VwdGlvbjEiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICJJbnRlcmFjdGlvbiBIb3Jtb25hbCBDb250cmFjcGV0aW9uIGFuZCBDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgY29uZGl0aW9uKSkpLA0KICAgICAgICAgY29uZGl0aW9uID0gaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9hZ2UiLCAiQWdlIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVldXJvXzUwMF8xMDAwIiwgIjUwMC0xMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fMTAwMF8yMDAwIiwgIjEwMDAtMjAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVldXJvXzIwMDBfMzAwMCIsICIyMDAwLTMwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb19ndF8zMDAwIiwgIj4zMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWRvbnRfdGVsbCIsICJkbyBub3QgdGVsbCIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG8yOG1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICIxMy0yOCBtb250aHMiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvNTJtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAiMjktNTIgbW9udGhzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfbW9yZXRoYW41Mm1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICI+NTIgbW9udGhzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2VkdWNhdGlvbl95ZWFycyIsICJZZWFycyBvZiBFZHVjYXRpb24iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX2V4dHJhIiwgIkV4dHJhdmVyc2lvbiIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfbmV1cm8iLCAiTmV1cm90aWNpc20iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX2FncmVlIiwgIkFncmVlYWJsZW5lc3MiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX2NvbnNjIiwgIkNvbnNjaWVudGlvdXNuZXNzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9vcGVuIiwgIk9wZW5uZXNzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX3JlbGlnaW9zaXR5IiwgIlJlbGlnaW9zaXR5IiwNCiAgICAgICAgICAgICAgICAgICAgICAgY29uZGl0aW9uKSkpKSkpKSkpKSkpKSkpKSwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGlzLm5hKGdyb3VwKSwgY29uZGl0aW9uLCBncm91cCksDQogICAgICAgICBjb25kaXRpb24gPSBmYWN0b3IoY29uZGl0aW9uLCBsZXZlbHMgPSByZXYoYygiSG9ybW9uYWwgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbmdydWVudCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiSW50ZXJhY3Rpb24gSG9ybW9uYWwgQ29udHJhY3BldGlvbiBhbmQgQ29uZ3J1ZW50IENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJBZ2UiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICI1MDAtMTAwMCBFdXJvIiwgIjEwMDAtMjAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiMjAwMC0zMDAwIEV1cm8iLCAiPjMwMDAgRXVybyIsICJkbyBub3QgdGVsbCIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIjEzLTI4IG1vbnRocyIsICIyOS01MiBtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICI+NTIgbW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiWWVhcnMgb2YgRWR1Y2F0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiRXh0cmF2ZXJzaW9uIiwgIk5ldXJvdGljaXNtIiwgIkFncmVlYWJsZW5lc3MiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiQ29uc2NpZW50aW91c25lc3MiLCJPcGVubmVzcyIsIlJlbGlnaW9zaXR5IikpKSwNCiAgICAgICAgIGdyb3VwID0gZmFjdG9yKGdyb3VwLCBsZXZlbHMgPSBjKCJDb250cmFjZXB0aW9uIiwgIkFnZSIsICJJbmNvbWUiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIlJlbGF0aW9uc2hpcCBEdXJhdGlvbiIsIlllYXJzIG9mIEVkdWNhdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiRXh0cmF2ZXJzaW9uIiwgIk5ldXJvdGljaXNtIiwgIkFncmVlYWJsZW5lc3MiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiQ29uc2NpZW50aW91c25lc3MiLCJPcGVubmVzcyIsIlJlbGlnaW9zaXR5IikpKSAlPiUNCiAgZ2dwbG90KGFlcyh5ID0gY29uZGl0aW9uLA0KICAgICAgICAgICAgIHggPSBjb25kaXRpb25fbWVhbiwNCiAgICAgICAgICAgICBmaWxsID0gc3RhdChhYnMoeCkgPCAwLjExKSkpICsNCiAgc3RhdF9oYWxmZXllKCkgKw0KICBnZW9tX3ZsaW5lKHhpbnRlcmNlcHQgPSBjKC0wLjExLCAwLjExKSwgbGluZXR5cGUgPSAiZG90dGVkIikgKw0KICBhcGF0aGVtZSArDQogIHRoZW1lKGxlZ2VuZC5wb3NpdGlvbiA9ICJub25lIikgKw0KICBzY2FsZV9maWxsX21hbnVhbCh2YWx1ZXMgPSBjKCJncmF5ODAiLCAic2t5Ymx1ZSIpKSArDQogIGxhYnMoeCA9ICJFZmZlY3QgU2l6ZSBFc3RpbWF0ZXMiLCB5ID0gIlByZWRpY3RvcnMiKQ0KYGBgDQoNCiMjIyBMaWJpZG8gey50YWJzZXR9DQojIyMjIE1vZGVsDQpgYGB7cn0NCm1fY29uZ3J1ZW5jeV9saWJpZG9fY29udHJvbGxlZCA9IGJybShkaWFyeV9saWJpZG9fbWVhbiB+DQogICAgICAgICAgICAgICAgICAgICAgICAgIGNvbnRyYWNlcHRpb25faG9ybW9uYWwgKiBjb25ncnVlbnRfY29udHJhY2VwdGlvbiArDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgIGFnZSArIG5ldF9pbmNvbWUgKyByZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIGVkdWNhdGlvbl95ZWFycyArDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICBiZmlfZXh0cmEgKyBiZmlfbmV1cm8gKyBiZmlfYWdyZWUgKyBiZmlfY29uc2MgKyBiZmlfb3BlbiArDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICByZWxpZ2lvc2l0eSwNCiAgICAgICAgICAgICAgICBkYXRhID0gZGF0YSwgZmFtaWx5ID0gZ2F1c3NpYW4oKSwNCiAgICAgICAgICAgICAgICBmaWxlID0gIm1fY29uZ3J1ZW5jeV9saWJpZG9fY29udHJvbGxlZCIpDQoNCmBgYA0KDQojIyMjIFN1bW1hcnkNCmBgYHtyfQ0Kc3VtbWFyeShtX2NvbmdydWVuY3lfbGliaWRvX2NvbnRyb2xsZWQsIGludGVydmFscyA9IFQsIHByb2IgPSAwLjkwKQ0KYGBgDQoNCiMjIyMgQ29tcGFyaXNvbiB3aXRoIFJPUEUNCmBgYHtyfQ0KcGxvdChlcXVpdmFsZW5jZV90ZXN0KG1fY29uZ3J1ZW5jeV9saWJpZG9fY29udHJvbGxlZCwgcmFuZ2UgPSBjKC0wLjA2LCAwLjA2KSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpKQ0KZXF1aXZhbGVuY2VfdGVzdChtX2NvbmdydWVuY3lfbGliaWRvX2NvbnRyb2xsZWQsIHJhbmdlID0gYygtMC4wNiwgMC4wNiksIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKQ0KYGBgDQoNCiMjIyMgUGxvdHMNCmBgYHtyIHdhcm5pbmcgPSBGQUxTRX0NCmNvbmRpdGlvbmFsX2VmZmVjdHMobV9jb25ncnVlbmN5X2xpYmlkb19jb250cm9sbGVkLCBhc2sgPSBGQUxTRSkNCmBgYA0KDQojIyMjIEZvcmVzdCBQbG90IGZvciBFZmZlY3QgU2l6ZXMgey5hY3RpdmV9DQpgYGB7cn0NCm1fY29uZ3J1ZW5jeV9saWJpZG9fY29udHJvbGxlZCAlPiUNCiAgDQogIHNwcmVhZF9kcmF3cyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMsIGJfY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xLA0KICAgICAgICAgICAgICAgYGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllczpjb25ncnVlbnRfY29udHJhY2VwdGlvbjFgLA0KICAgICAgICAgICAgICAgYl9hZ2UsDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzUwMF8xMDAwLCBiX25ldF9pbmNvbWVldXJvXzEwMDBfMjAwMCwgDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzIwMDBfMzAwMCwgYl9uZXRfaW5jb21lZXVyb19ndF8zMDAwLCBiX25ldF9pbmNvbWVkb250X3RlbGwsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzI4bW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG81Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF9tb3JldGhhbjUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9lZHVjYXRpb25feWVhcnMsDQogICAgICAgICAgICAgICBiX2JmaV9leHRyYSwgYl9iZmlfbmV1cm8sIGJfYmZpX2FncmVlLCBiX2JmaV9jb25zYywgYl9iZmlfb3BlbiwgDQogICAgICAgICAgICAgICBiX3JlbGlnaW9zaXR5KSAlPiUNCiAgcGl2b3RfbG9uZ2VyKGNvbHMgPSBjKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcywgYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEsDQogICAgICAgICAgICAgICBgYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzOmNvbmdydWVudF9jb250cmFjZXB0aW9uMWAsDQogICAgICAgICAgICAgICAgICAgICAgICBiX2FnZSwNCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fNTAwXzEwMDAsIGJfbmV0X2luY29tZWV1cm9fMTAwMF8yMDAwLCANCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fMjAwMF8zMDAwLCBiX25ldF9pbmNvbWVldXJvX2d0XzMwMDAsIGJfbmV0X2luY29tZWRvbnRfdGVsbCwNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvMjhtb250aHMsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX21vcmV0aGFuNTJtb250aHMsDQogICAgICAgICAgICAgICBiX2VkdWNhdGlvbl95ZWFycywNCiAgICAgICAgICAgICAgIGJfYmZpX2V4dHJhLCBiX2JmaV9uZXVybywgYl9iZmlfYWdyZWUsIGJfYmZpX2NvbnNjLCBiX2JmaV9vcGVuLCANCiAgICAgICAgICAgICAgIGJfcmVsaWdpb3NpdHkpLA0KICAgICAgICAgICAgICAgbmFtZXNfdG8gPSAiY29uZGl0aW9uIiwNCiAgICAgICAgICAgICAgIHZhbHVlc190byA9ICJyX2NvbmRpdGlvbiIpICU+JQ0KICBtdXRhdGUoY29uZGl0aW9uX21lYW4gPSByX2NvbmRpdGlvbiwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3IiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIlJlbGF0aW9uc2hpcCBEdXJhdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfbmV0X2luY29tZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkluY29tZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgTkEpKSwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnRyYWNlcHRpb24iLCBncm91cCksDQogICAgICAgICBjb25kaXRpb24gPSBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXM6Y29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAiSW50ZXJhY3Rpb24gSG9ybW9uYWwgQ29udHJhY3BldGlvbiBhbmQgQ29uZ3J1ZW50IENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgIGNvbmRpdGlvbikpKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGlmZWxzZShjb25kaXRpb24gPT0gImJfYWdlIiwgIkFnZSIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb181MDBfMTAwMCIsICI1MDAtMTAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVldXJvXzEwMDBfMjAwMCIsICIxMDAwLTIwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb18yMDAwXzMwMDAiLCAiMjAwMC0zMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fZ3RfMzAwMCIsICI+MzAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVkb250X3RlbGwiLCAiZG8gbm90IHRlbGwiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvMjhtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAiMTMtMjggbW9udGhzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzUybW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIjI5LTUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX21vcmV0aGFuNTJtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAiPjUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9lZHVjYXRpb25feWVhcnMiLCAiWWVhcnMgb2YgRWR1Y2F0aW9uIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9leHRyYSIsICJFeHRyYXZlcnNpb24iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX25ldXJvIiwgIk5ldXJvdGljaXNtIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9hZ3JlZSIsICJBZ3JlZWFibGVuZXNzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9jb25zYyIsICJDb25zY2llbnRpb3VzbmVzcyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfb3BlbiIsICJPcGVubmVzcyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxpZ2lvc2l0eSIsICJSZWxpZ2lvc2l0eSIsDQogICAgICAgICAgICAgICAgICAgICAgIGNvbmRpdGlvbikpKSkpKSkpKSkpKSkpKSksDQogICAgICAgICBncm91cCA9IGlmZWxzZShpcy5uYShncm91cCksIGNvbmRpdGlvbiwgZ3JvdXApLA0KICAgICAgICAgY29uZGl0aW9uID0gZmFjdG9yKGNvbmRpdGlvbiwgbGV2ZWxzID0gcmV2KGMoIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkludGVyYWN0aW9uIEhvcm1vbmFsIENvbnRyYWNwZXRpb24gYW5kIENvbmdydWVudCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiQWdlIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiNTAwLTEwMDAgRXVybyIsICIxMDAwLTIwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIjIwMDAtMzAwMCBFdXJvIiwgIj4zMDAwIEV1cm8iLCAiZG8gbm90IHRlbGwiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICIxMy0yOCBtb250aHMiLCAiMjktNTIgbW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiPjUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIlllYXJzIG9mIEVkdWNhdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkV4dHJhdmVyc2lvbiIsICJOZXVyb3RpY2lzbSIsICJBZ3JlZWFibGVuZXNzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnNjaWVudGlvdXNuZXNzIiwiT3Blbm5lc3MiLCJSZWxpZ2lvc2l0eSIpKSksDQogICAgICAgICBncm91cCA9IGZhY3Rvcihncm91cCwgbGV2ZWxzID0gYygiQ29udHJhY2VwdGlvbiIsICJBZ2UiLCAiSW5jb21lIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJSZWxhdGlvbnNoaXAgRHVyYXRpb24iLCJZZWFycyBvZiBFZHVjYXRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkV4dHJhdmVyc2lvbiIsICJOZXVyb3RpY2lzbSIsICJBZ3JlZWFibGVuZXNzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnNjaWVudGlvdXNuZXNzIiwiT3Blbm5lc3MiLCJSZWxpZ2lvc2l0eSIpKSkgJT4lDQogIGdncGxvdChhZXMoeSA9IGNvbmRpdGlvbiwNCiAgICAgICAgICAgICB4ID0gY29uZGl0aW9uX21lYW4sDQogICAgICAgICAgICAgZmlsbCA9IHN0YXQoYWJzKHgpIDwgMC4wNikpKSArDQogIHN0YXRfaGFsZmV5ZSgpICsNCiAgZ2VvbV92bGluZSh4aW50ZXJjZXB0ID0gYygtMC4wNiwgMC4wNiksIGxpbmV0eXBlID0gImRvdHRlZCIpICsNCiAgYXBhdGhlbWUgKw0KICB0aGVtZShsZWdlbmQucG9zaXRpb24gPSAibm9uZSIpICsNCiAgc2NhbGVfZmlsbF9tYW51YWwodmFsdWVzID0gYygiZ3JheTgwIiwgInNreWJsdWUiKSkgKw0KICBsYWJzKHggPSAiRWZmZWN0IFNpemUgRXN0aW1hdGVzIiwgeSA9ICJQcmVkaWN0b3JzIikNCmBgYA0KDQoNCiMjIyBTZXh1YWwgRnJlcXVlbmN5IChQZW5ldHJhdGl2ZSBJbnRlcmNvdXJzZSkgey50YWJzZXR9DQojIyMjIE1vZGVsDQpgYGB7cn0NCm1fY29uZ3J1ZW5jeV9zZXhmcmVxcGVuX2NvbnRyb2xsZWQgPSBicm0oZGlhcnlfc2V4X2FjdGl2ZV9zZXhfc3VtIH4NCiAgICAgICAgICAgICAgICAgICAgICAgIG9mZnNldChsb2cobnVtYmVyX29mX2RheXMpKSArDQogICAgICAgICAgICAgICAgICAgICAgICBjb250cmFjZXB0aW9uX2hvcm1vbmFsICogY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24gKw0KICAgICAgICAgICAgICAgICAgICAgICAgYWdlICsgbmV0X2luY29tZSArIHJlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3IgKw0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgZWR1Y2F0aW9uX3llYXJzICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIGJmaV9leHRyYSArIGJmaV9uZXVybyArIGJmaV9hZ3JlZSArIGJmaV9jb25zYyArIGJmaV9vcGVuICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIHJlbGlnaW9zaXR5LA0KICAgICAgICAgICAgICAgIGRhdGEgPSBkYXRhLCBmYW1pbHkgPSBwb2lzc29uKCksDQogICAgICAgICAgICAgICAgZmlsZSA9ICJtX2NvbmdydWVuY3lfc2V4ZnJlcXBlbl9jb250cm9sbGVkIikNCmBgYA0KDQojIyMjIFN1bW1hcnkNCmBgYHtyfQ0Kc3VtbWFyeShtX2NvbmdydWVuY3lfc2V4ZnJlcXBlbl9jb250cm9sbGVkLCBpbnRlcnZhbHMgPSBULCBwcm9iID0gMC45MCkNCmBgYA0KDQojIyMjIENvbXBhcmlzb24gd2l0aCBST1BFDQpgYGB7cn0NCnBsb3QoZXF1aXZhbGVuY2VfdGVzdChtX2NvbmdydWVuY3lfc2V4ZnJlcXBlbl9jb250cm9sbGVkLCByYW5nZSA9IGMoLTAuMDUsIDAuMDUpLA0KICAgICAgICAgICAgICAgICAgICAgIGNpID0gMC45MCwNCiAgICAgICAgICAgICAgICAgICAgICBwYXJhbWV0ZXJzID0gImNvbnRyYWNlcHRpb24iKSkNCmVxdWl2YWxlbmNlX3Rlc3QobV9jb25ncnVlbmN5X3NleGZyZXFwZW5fY29udHJvbGxlZCwgcmFuZ2UgPSBjKC0wLjA1LCAwLjA1KSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpDQpgYGANCg0KIyMjIyBQbG90cw0KYGBge3Igd2FybmluZyA9IEZBTFNFfQ0KY29uZGl0aW9uYWxfZWZmZWN0cyhtX2NvbmdydWVuY3lfc2V4ZnJlcXBlbl9jb250cm9sbGVkLA0KICAgICAgICAgICAgICAgICAgICBlZmZlY3RzID0gImNvbnRyYWNlcHRpb25faG9ybW9uYWw6Y29uZ3J1ZW50X2NvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICBjb25kaXRpb25zID0gZGF0YS5mcmFtZShudW1iZXJfb2ZfZGF5cyA9IDEpKQ0KYGBgDQoNCiMjIyMgRm9yZXN0IFBsb3QgZm9yIEVmZmVjdCBTaXplcyB7LmFjdGl2ZX0NCmBgYHtyfQ0KbV9jb25ncnVlbmN5X3NleGZyZXFwZW5fY29udHJvbGxlZCAlPiUNCg0KICBzcHJlYWRfZHJhd3MoYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzLCBiX2NvbmdydWVudF9jb250cmFjZXB0aW9uMSwNCiAgICAgICAgICAgICAgIGBiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXM6Y29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xYCwNCiAgICAgICAgICAgICAgIGJfYWdlLA0KICAgICAgICAgICAgICAgYl9uZXRfaW5jb21lZXVyb181MDBfMTAwMCwgYl9uZXRfaW5jb21lZXVyb18xMDAwXzIwMDAsIA0KICAgICAgICAgICAgICAgYl9uZXRfaW5jb21lZXVyb18yMDAwXzMwMDAsIGJfbmV0X2luY29tZWV1cm9fZ3RfMzAwMCwgYl9uZXRfaW5jb21lZG9udF90ZWxsLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG8yOG1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvNTJtb250aHMsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfbW9yZXRoYW41Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfZWR1Y2F0aW9uX3llYXJzLA0KICAgICAgICAgICAgICAgYl9iZmlfZXh0cmEsIGJfYmZpX25ldXJvLCBiX2JmaV9hZ3JlZSwgYl9iZmlfY29uc2MsIGJfYmZpX29wZW4sIA0KICAgICAgICAgICAgICAgYl9yZWxpZ2lvc2l0eSkgJT4lDQogIHBpdm90X2xvbmdlcihjb2xzID0gYyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMsIGJfY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xLA0KICAgICAgICAgICAgICAgYGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllczpjb25ncnVlbnRfY29udHJhY2VwdGlvbjFgLA0KICAgICAgICAgICAgICAgICAgICAgICAgYl9hZ2UsDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzUwMF8xMDAwLCBiX25ldF9pbmNvbWVldXJvXzEwMDBfMjAwMCwgDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzIwMDBfMzAwMCwgYl9uZXRfaW5jb21lZXVyb19ndF8zMDAwLCBiX25ldF9pbmNvbWVkb250X3RlbGwsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzI4bW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG81Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF9tb3JldGhhbjUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9lZHVjYXRpb25feWVhcnMsDQogICAgICAgICAgICAgICBiX2JmaV9leHRyYSwgYl9iZmlfbmV1cm8sIGJfYmZpX2FncmVlLCBiX2JmaV9jb25zYywgYl9iZmlfb3BlbiwgDQogICAgICAgICAgICAgICBiX3JlbGlnaW9zaXR5KSwNCiAgICAgICAgICAgICAgIG5hbWVzX3RvID0gImNvbmRpdGlvbiIsDQogICAgICAgICAgICAgICB2YWx1ZXNfdG8gPSAicl9jb25kaXRpb24iKSAlPiUNCiAgbXV0YXRlKGNvbmRpdGlvbl9tZWFuID0gcl9jb25kaXRpb24sDQogICAgICAgICBncm91cCA9IGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJSZWxhdGlvbnNoaXAgRHVyYXRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX25ldF9pbmNvbWUiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJJbmNvbWUiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIE5BKSksDQogICAgICAgICBncm91cCA9IGlmZWxzZShjb25kaXRpb24gJWNvbnRhaW5zJSAib250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJDb250cmFjZXB0aW9uIiwgZ3JvdXApLA0KICAgICAgICAgY29uZGl0aW9uID0gaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICJIb3Jtb25hbCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAiQ29uZ3J1ZW50IENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzOmNvbmdydWVudF9jb250cmFjZXB0aW9uMSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgIkludGVyYWN0aW9uIEhvcm1vbmFsIENvbnRyYWNwZXRpb24gYW5kIENvbmdydWVudCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICBjb25kaXRpb24pKSksDQogICAgICAgICBjb25kaXRpb24gPSBpZmVsc2UoY29uZGl0aW9uID09ICJiX2FnZSIsICJBZ2UiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fNTAwXzEwMDAiLCAiNTAwLTEwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb18xMDAwXzIwMDAiLCAiMTAwMC0yMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fMjAwMF8zMDAwIiwgIjIwMDAtMzAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVldXJvX2d0XzMwMDAiLCAiPjMwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZG9udF90ZWxsIiwgImRvIG5vdCB0ZWxsIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzI4bW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIjEzLTI4IG1vbnRocyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG81Mm1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICIyOS01MiBtb250aHMiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF9tb3JldGhhbjUybW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIj41MiBtb250aHMiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfZWR1Y2F0aW9uX3llYXJzIiwgIlllYXJzIG9mIEVkdWNhdGlvbiIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfZXh0cmEiLCAiRXh0cmF2ZXJzaW9uIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9uZXVybyIsICJOZXVyb3RpY2lzbSIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfYWdyZWUiLCAiQWdyZWVhYmxlbmVzcyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfY29uc2MiLCAiQ29uc2NpZW50aW91c25lc3MiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX29wZW4iLCAiT3Blbm5lc3MiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsaWdpb3NpdHkiLCAiUmVsaWdpb3NpdHkiLA0KICAgICAgICAgICAgICAgICAgICAgICBjb25kaXRpb24pKSkpKSkpKSkpKSkpKSkpLA0KICAgICAgICAgZ3JvdXAgPSBpZmVsc2UoaXMubmEoZ3JvdXApLCBjb25kaXRpb24sIGdyb3VwKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGZhY3Rvcihjb25kaXRpb24sIGxldmVscyA9IHJldihjKCJIb3Jtb25hbCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiQ29uZ3J1ZW50IENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJJbnRlcmFjdGlvbiBIb3Jtb25hbCBDb250cmFjcGV0aW9uIGFuZCBDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkFnZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIjUwMC0xMDAwIEV1cm8iLCAiMTAwMC0yMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICIyMDAwLTMwMDAgRXVybyIsICI+MzAwMCBFdXJvIiwgImRvIG5vdCB0ZWxsIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiMTMtMjggbW9udGhzIiwgIjI5LTUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIj41MiBtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJZZWFycyBvZiBFZHVjYXRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJFeHRyYXZlcnNpb24iLCAiTmV1cm90aWNpc20iLCAiQWdyZWVhYmxlbmVzcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25zY2llbnRpb3VzbmVzcyIsIk9wZW5uZXNzIiwiUmVsaWdpb3NpdHkiKSkpLA0KICAgICAgICAgZ3JvdXAgPSBmYWN0b3IoZ3JvdXAsIGxldmVscyA9IGMoIkNvbnRyYWNlcHRpb24iLCAiQWdlIiwgIkluY29tZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiUmVsYXRpb25zaGlwIER1cmF0aW9uIiwiWWVhcnMgb2YgRWR1Y2F0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJFeHRyYXZlcnNpb24iLCAiTmV1cm90aWNpc20iLCAiQWdyZWVhYmxlbmVzcyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25zY2llbnRpb3VzbmVzcyIsIk9wZW5uZXNzIiwiUmVsaWdpb3NpdHkiKSkpICU+JQ0KICBnZ3Bsb3QoYWVzKHkgPSBjb25kaXRpb24sDQogICAgICAgICAgICAgeCA9IGNvbmRpdGlvbl9tZWFuLA0KICAgICAgICAgICAgIGZpbGwgPSBzdGF0KGFicyh4KSA8IDAuMDUpKSkgKw0KICBzdGF0X2hhbGZleWUoKSArDQogIGdlb21fdmxpbmUoeGludGVyY2VwdCA9IGMoLTAuMDUsIDAuMDUpLCBsaW5ldHlwZSA9ICJkb3R0ZWQiKSArDQogIGFwYXRoZW1lICsNCiAgdGhlbWUobGVnZW5kLnBvc2l0aW9uID0gIm5vbmUiKSArDQogIHNjYWxlX2ZpbGxfbWFudWFsKHZhbHVlcyA9IGMoImdyYXk4MCIsICJza3libHVlIikpICsNCiAgbGFicyh4ID0gIkVmZmVjdCBTaXplIEVzdGltYXRlcyIsIHkgPSAiUHJlZGljdG9ycyIpDQpgYGANCg0KIyMjIE1hc3R1cmJhdGlvbiBGcmVxdWVuY3kgey50YWJzZXR9DQojIyMjIE1vZGVsDQpgYGB7cn0NCm1fY29uZ3J1ZW5jeV9tYXNmcmVxX2NvbnRyb2xsZWQgPSBicm0oZGlhcnlfbWFzdHVyYmF0aW9uX3N1bSB+DQogICAgICAgICAgICAgICAgICAgICAgICBvZmZzZXQobG9nKG51bWJlcl9vZl9kYXlzKSkgKw0KICAgICAgICAgICAgICAgICAgICAgICAgY29udHJhY2VwdGlvbl9ob3Jtb25hbCAqIGNvbmdydWVudF9jb250cmFjZXB0aW9uICsNCiAgICAgICAgICAgICAgICAgICAgICAgIGFnZSArIG5ldF9pbmNvbWUgKyByZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yICsNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIGVkdWNhdGlvbl95ZWFycyArDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICBiZmlfZXh0cmEgKyBiZmlfbmV1cm8gKyBiZmlfYWdyZWUgKyBiZmlfY29uc2MgKyBiZmlfb3BlbiArDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICByZWxpZ2lvc2l0eSwNCiAgICAgICAgICAgICAgICBkYXRhID0gZGF0YSwgZmFtaWx5ID0gcG9pc3NvbigpLA0KICAgICAgICAgICAgICAgIGZpbGUgPSAibV9jb25ncnVlbmN5X21hc2ZyZXFfY29udHJvbGxlZCIpDQpgYGANCg0KIyMjIyBTdW1tYXJ5DQpgYGB7cn0NCnN1bW1hcnkobV9jb25ncnVlbmN5X21hc2ZyZXFfY29udHJvbGxlZCwgaW50ZXJ2YWxzID0gVCwgcHJvYiA9IDAuOTApDQpgYGANCg0KIyMjIyBDb21wYXJpc29uIHdpdGggUk9QRQ0KYGBge3J9DQpwbG90KGVxdWl2YWxlbmNlX3Rlc3QobV9jb25ncnVlbmN5X21hc2ZyZXFfY29udHJvbGxlZCwgcmFuZ2UgPSBjKC0wLjA1LCAwLjA1KSwgY2kgPSAwLjkwLA0KICAgICAgICAgICAgICAgICAgICAgIHBhcmFtZXRlcnMgPSAiY29udHJhY2VwdGlvbiIpKQ0KZXF1aXZhbGVuY2VfdGVzdChtX2NvbmdydWVuY3lfbWFzZnJlcV9jb250cm9sbGVkLCByYW5nZSA9IGMoLTAuMDUsIDAuMDUpLCBjaSA9IDAuOTAsDQogICAgICAgICAgICAgICAgICAgICAgcGFyYW1ldGVycyA9ICJjb250cmFjZXB0aW9uIikNCmBgYA0KDQojIyMjIFBsb3RzDQpgYGB7ciB3YXJuaW5nID0gRkFMU0V9DQpjb25kaXRpb25hbF9lZmZlY3RzKG1fY29uZ3J1ZW5jeV9tYXNmcmVxX2NvbnRyb2xsZWQsDQogICAgICAgICAgICAgICAgICAgIGVmZmVjdHMgPSAiY29udHJhY2VwdGlvbl9ob3Jtb25hbDpjb25ncnVlbnRfY29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgIGNvbmRpdGlvbnMgPSBkYXRhLmZyYW1lKG51bWJlcl9vZl9kYXlzID0gMSkpDQpgYGANCg0KIyMjIyBGb3Jlc3QgUGxvdCBmb3IgRWZmZWN0IFNpemVzIHsuYWN0aXZlfQ0KYGBge3J9DQptX2NvbmdydWVuY3lfbWFzZnJlcV9jb250cm9sbGVkICU+JQ0KDQogIHNwcmVhZF9kcmF3cyhiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMsIGJfY29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xLA0KICAgICAgICAgICAgICAgYGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllczpjb25ncnVlbnRfY29udHJhY2VwdGlvbjFgLA0KICAgICAgICAgICAgICAgYl9hZ2UsDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzUwMF8xMDAwLCBiX25ldF9pbmNvbWVldXJvXzEwMDBfMjAwMCwgDQogICAgICAgICAgICAgICBiX25ldF9pbmNvbWVldXJvXzIwMDBfMzAwMCwgYl9uZXRfaW5jb21lZXVyb19ndF8zMDAwLCBiX25ldF9pbmNvbWVkb250X3RlbGwsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzI4bW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX3VwdG81Mm1vbnRocywNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF9tb3JldGhhbjUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9lZHVjYXRpb25feWVhcnMsDQogICAgICAgICAgICAgICBiX2JmaV9leHRyYSwgYl9iZmlfbmV1cm8sIGJfYmZpX2FncmVlLCBiX2JmaV9jb25zYywgYl9iZmlfb3BlbiwgDQogICAgICAgICAgICAgICBiX3JlbGlnaW9zaXR5KSAlPiUNCiAgcGl2b3RfbG9uZ2VyKGNvbHMgPSBjKGJfY29udHJhY2VwdGlvbl9ob3Jtb25hbHllcywgYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEsDQogICAgICAgICAgICAgICBgYl9jb250cmFjZXB0aW9uX2hvcm1vbmFseWVzOmNvbmdydWVudF9jb250cmFjZXB0aW9uMWAsDQogICAgICAgICAgICAgICAgICAgICAgICBiX2FnZSwNCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fNTAwXzEwMDAsIGJfbmV0X2luY29tZWV1cm9fMTAwMF8yMDAwLCANCiAgICAgICAgICAgICAgIGJfbmV0X2luY29tZWV1cm9fMjAwMF8zMDAwLCBiX25ldF9pbmNvbWVldXJvX2d0XzMwMDAsIGJfbmV0X2luY29tZWRvbnRfdGVsbCwNCiAgICAgICAgICAgICAgIGJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvMjhtb250aHMsDQogICAgICAgICAgICAgICBiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzUybW9udGhzLA0KICAgICAgICAgICAgICAgYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX21vcmV0aGFuNTJtb250aHMsDQogICAgICAgICAgICAgICBiX2VkdWNhdGlvbl95ZWFycywNCiAgICAgICAgICAgICAgIGJfYmZpX2V4dHJhLCBiX2JmaV9uZXVybywgYl9iZmlfYWdyZWUsIGJfYmZpX2NvbnNjLCBiX2JmaV9vcGVuLCANCiAgICAgICAgICAgICAgIGJfcmVsaWdpb3NpdHkpLA0KICAgICAgICAgICAgICAgbmFtZXNfdG8gPSAiY29uZGl0aW9uIiwNCiAgICAgICAgICAgICAgIHZhbHVlc190byA9ICJyX2NvbmRpdGlvbiIpICU+JQ0KICBtdXRhdGUoY29uZGl0aW9uX21lYW4gPSByX2NvbmRpdGlvbiwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3IiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIlJlbGF0aW9uc2hpcCBEdXJhdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uICVjb250YWlucyUgImJfbmV0X2luY29tZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkluY29tZSIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgTkEpKSwNCiAgICAgICAgIGdyb3VwID0gaWZlbHNlKGNvbmRpdGlvbiAlY29udGFpbnMlICJvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnRyYWNlcHRpb24iLCBncm91cCksDQogICAgICAgICBjb25kaXRpb24gPSBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXMiLA0KICAgICAgICAgICAgICAgICAgICAgICAgIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9jb25ncnVlbnRfY29udHJhY2VwdGlvbjEiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2NvbnRyYWNlcHRpb25faG9ybW9uYWx5ZXM6Y29uZ3J1ZW50X2NvbnRyYWNlcHRpb24xIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAiSW50ZXJhY3Rpb24gSG9ybW9uYWwgQ29udHJhY3BldGlvbiBhbmQgQ29uZ3J1ZW50IENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgIGNvbmRpdGlvbikpKSwNCiAgICAgICAgIGNvbmRpdGlvbiA9IGlmZWxzZShjb25kaXRpb24gPT0gImJfYWdlIiwgIkFnZSIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb181MDBfMTAwMCIsICI1MDAtMTAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVldXJvXzEwMDBfMjAwMCIsICIxMDAwLTIwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9uZXRfaW5jb21lZXVyb18yMDAwXzMwMDAiLCAiMjAwMC0zMDAwIEV1cm8iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfbmV0X2luY29tZWV1cm9fZ3RfMzAwMCIsICI+MzAwMCBFdXJvIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX25ldF9pbmNvbWVkb250X3RlbGwiLCAiZG8gbm90IHRlbGwiLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfcmVsYXRpb25zaGlwX2R1cmF0aW9uX2ZhY3RvclBhcnRuZXJlZF91cHRvMjhtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAiMTMtMjggbW9udGhzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX3JlbGF0aW9uc2hpcF9kdXJhdGlvbl9mYWN0b3JQYXJ0bmVyZWRfdXB0bzUybW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgIjI5LTUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxhdGlvbnNoaXBfZHVyYXRpb25fZmFjdG9yUGFydG5lcmVkX21vcmV0aGFuNTJtb250aHMiLA0KICAgICAgICAgICAgICAgICAgICAgICAiPjUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9lZHVjYXRpb25feWVhcnMiLCAiWWVhcnMgb2YgRWR1Y2F0aW9uIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9leHRyYSIsICJFeHRyYXZlcnNpb24iLA0KICAgICAgICAgICAgICAgIGlmZWxzZShjb25kaXRpb24gPT0gImJfYmZpX25ldXJvIiwgIk5ldXJvdGljaXNtIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9hZ3JlZSIsICJBZ3JlZWFibGVuZXNzIiwNCiAgICAgICAgICAgICAgICBpZmVsc2UoY29uZGl0aW9uID09ICJiX2JmaV9jb25zYyIsICJDb25zY2llbnRpb3VzbmVzcyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9iZmlfb3BlbiIsICJPcGVubmVzcyIsDQogICAgICAgICAgICAgICAgaWZlbHNlKGNvbmRpdGlvbiA9PSAiYl9yZWxpZ2lvc2l0eSIsICJSZWxpZ2lvc2l0eSIsDQogICAgICAgICAgICAgICAgICAgICAgIGNvbmRpdGlvbikpKSkpKSkpKSkpKSkpKSksDQogICAgICAgICBncm91cCA9IGlmZWxzZShpcy5uYShncm91cCksIGNvbmRpdGlvbiwgZ3JvdXApLA0KICAgICAgICAgY29uZGl0aW9uID0gZmFjdG9yKGNvbmRpdGlvbiwgbGV2ZWxzID0gcmV2KGMoIkhvcm1vbmFsIENvbnRyYWNlcHRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJDb25ncnVlbnQgQ29udHJhY2VwdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkludGVyYWN0aW9uIEhvcm1vbmFsIENvbnRyYWNwZXRpb24gYW5kIENvbmdydWVudCBDb250cmFjZXB0aW9uIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiQWdlIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiNTAwLTEwMDAgRXVybyIsICIxMDAwLTIwMDAgRXVybyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIjIwMDAtMzAwMCBFdXJvIiwgIj4zMDAwIEV1cm8iLCAiZG8gbm90IHRlbGwiLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICIxMy0yOCBtb250aHMiLCAiMjktNTIgbW9udGhzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAiPjUyIG1vbnRocyIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIlllYXJzIG9mIEVkdWNhdGlvbiIsDQogICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkV4dHJhdmVyc2lvbiIsICJOZXVyb3RpY2lzbSIsICJBZ3JlZWFibGVuZXNzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnNjaWVudGlvdXNuZXNzIiwiT3Blbm5lc3MiLCJSZWxpZ2lvc2l0eSIpKSksDQogICAgICAgICBncm91cCA9IGZhY3Rvcihncm91cCwgbGV2ZWxzID0gYygiQ29udHJhY2VwdGlvbiIsICJBZ2UiLCAiSW5jb21lIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICJSZWxhdGlvbnNoaXAgRHVyYXRpb24iLCJZZWFycyBvZiBFZHVjYXRpb24iLA0KICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkV4dHJhdmVyc2lvbiIsICJOZXVyb3RpY2lzbSIsICJBZ3JlZWFibGVuZXNzIiwNCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgIkNvbnNjaWVudGlvdXNuZXNzIiwiT3Blbm5lc3MiLCJSZWxpZ2lvc2l0eSIpKSkgJT4lDQogIGdncGxvdChhZXMoeSA9IGNvbmRpdGlvbiwNCiAgICAgICAgICAgICB4ID0gY29uZGl0aW9uX21lYW4sDQogICAgICAgICAgICAgZmlsbCA9IHN0YXQoYWJzKHgpIDwgMC4wNSkpKSArDQogIHN0YXRfaGFsZmV5ZSgpICsNCiAgZ2VvbV92bGluZSh4aW50ZXJjZXB0ID0gYygtMC4wNSwgMC4wNSksIGxpbmV0eXBlID0gImRvdHRlZCIpICsNCiAgYXBhdGhlbWUgKw0KICB0aGVtZShsZWdlbmQucG9zaXRpb24gPSAibm9uZSIpICsNCiAgc2NhbGVfZmlsbF9tYW51YWwodmFsdWVzID0gYygiZ3JheTgwIiwgInNreWJsdWUiKSkgKw0KICBsYWJzKHggPSAiRWZmZWN0IFNpemUgRXN0aW1hdGVzIiwgeSA9ICJQcmVkaWN0b3JzIikNCmBgYA0KDQojIyMgTW9kZWwgQ29tcGFyaXNvbg0KIyMjIyBBdHRyYWN0aXZlbmVzcyBvZiBwYXJ0bmVyDQpgYGB7cn0NCmNvbXBhcmVfbW9kZWxzID0gbG9vKG1faGNfYXRyciwgbV9jb25ncnVlbmN5X2F0cnIpDQpgYGANCkxvb0lDIGZpcnN0IG1vZGVsOiBgciByb3VuZChjb21wYXJlX21vZGVscyRsb29zJG1faGNfYXRyciRlc3RpbWF0ZXNbMywxXSwgMilgDQoNCkxvb0lDIHNlY29uZCBtb2RlbDogYHIgcm91bmQoY29tcGFyZV9tb2RlbHMkbG9vcyRtX2NvbmdydWVuY3lfYXRyciRlc3RpbWF0ZXNbMywxXSwgMilgDQoNCk1vZGVsIENvbXBhcmlzb25zOiBUaGUgZGlmZmVyZW5jZSBiZXR3ZWVuIG1vZGVscyBpcyBgciByb3VuZChjb21wYXJlX21vZGVscyRkaWZmc1syLDFdLCAyKWAgY29tcGFyZWQgdG8gYSBzdGFuZGFyZCBlcnJvciBvZiBgciByb3VuZChjb21wYXJlX21vZGVscyRkaWZmc1syLDJdLCAyKWANCmBgYHtyfQ0KY29tcGFyZV9tb2RlbHMNCmBgYA0KDQojIyMjIFJlbGF0aW9uc2hpcCBzYXRpc2ZhY3Rpb24NCmBgYHtyfQ0KY29tcGFyZV9tb2RlbHMgPSBsb28obV9oY19yZWxzYXQsIG1fY29uZ3J1ZW5jeV9yZWxzYXQpDQpgYGANCkxvb0lDIGZpcnN0IG1vZGVsOiBgciByb3VuZChjb21wYXJlX21vZGVscyRsb29zJG1faGNfcmVsc2F0JGVzdGltYXRlc1szLDFdLCAyKWANCg0KTG9vSUMgc2Vjb25kIG1vZGVsOiBgciByb3VuZChjb21wYXJlX21vZGVscyRsb29zJG1fY29uZ3J1ZW5jeV9yZWxzYXQkZXN0aW1hdGVzWzMsMV0sIDIpYA0KDQpNb2RlbCBDb21wYXJpc29uczogVGhlIGRpZmZlcmVuY2UgYmV0d2VlbiBtb2RlbHMgaXMgYHIgcm91bmQoY29tcGFyZV9tb2RlbHMkZGlmZnNbMiwxXSwgMilgIGNvbXBhcmVkIHRvIGEgc3RhbmRhcmQgZXJyb3Igb2YgYHIgcm91bmQoY29tcGFyZV9tb2RlbHMkZGlmZnNbMiwyXSwgMilgDQpgYGB7cn0NCmNvbXBhcmVfbW9kZWxzDQpgYGANCg0KIyMjIyBTZXh1YWwgU2F0aXNmYWN0aW9uDQpgYGB7cn0NCm1faGNfc2V4c2F0JGRhdGEkc2F0aXNmYWN0aW9uX3NleHVhbF9pbnRlcmNvdXJzZSA9DQogIGFzLm51bWVyaWMobV9oY19zZXhzYXQkZGF0YSRzYXRpc2ZhY3Rpb25fc2V4dWFsX2ludGVyY291cnNlKQ0KbV9jb25ncnVlbmN5X3NleHNhdCRkYXRhJHNhdGlzZmFjdGlvbl9zZXh1YWxfaW50ZXJjb3Vyc2UgPQ0KICBhcy5udW1lcmljKG1fY29uZ3J1ZW5jeV9zZXhzYXQkZGF0YSRzYXRpc2ZhY3Rpb25fc2V4dWFsX2ludGVyY291cnNlKQ0KDQpjb21wYXJlX21vZGVscyA9IGxvbyhtX2hjX3NleHNhdCwgbV9jb25ncnVlbmN5X3NleHNhdCkNCmBgYA0KTG9vSUMgZmlyc3QgbW9kZWw6IGByIHJvdW5kKGNvbXBhcmVfbW9kZWxzJGxvb3MkbV9oY19zZXhzYXQkZXN0aW1hdGVzWzMsMV0sIDIpYA0KDQpMb29JQyBzZWNvbmQgbW9kZWw6IGByIHJvdW5kKGNvbXBhcmVfbW9kZWxzJGxvb3MkbV9jb25ncnVlbmN5X3NleHNhdCRlc3RpbWF0ZXNbMywxXSwgMilgDQoNCk1vZGVsIENvbXBhcmlzb25zOiBUaGUgZGlmZmVyZW5jZSBiZXR3ZWVuIG1vZGVscyBpcyBgciByb3VuZChjb21wYXJlX21vZGVscyRkaWZmc1syLDFdLCAyKWAgY29tcGFyZWQgdG8gYSBzdGFuZGFyZCBlcnJvciBvZiBgciByb3VuZChjb21wYXJlX21vZGVscyRkaWZmc1syLDJdLCAyKWANCmBgYHtyfQ0KY29tcGFyZV9tb2RlbHMNCmBgYA0KDQoNCg==